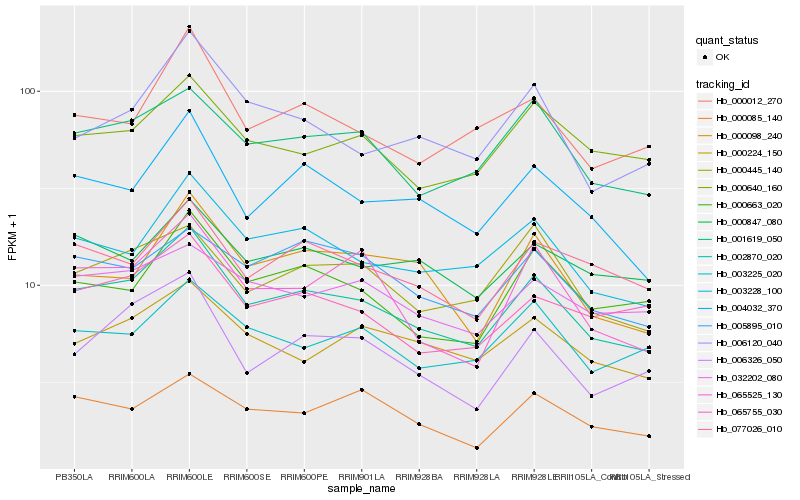
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002870_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003228_100 |
0.0722330693 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 3 |
Hb_004032_370 |
0.0764729995 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 4 |
Hb_001619_050 |
0.0764840608 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000663_020 |
0.0842308073 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_077026_010 |
0.0880217283 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 7 |
Hb_000445_140 |
0.0897051071 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 8 |
Hb_000012_270 |
0.0913696764 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 9 |
Hb_000098_240 |
0.092251425 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 10 |
Hb_000640_160 |
0.0924964082 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 11 |
Hb_000224_150 |
0.0929084471 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_005895_010 |
0.0931324495 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006326_050 |
0.0932132561 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_065755_030 |
0.0932414347 |
- |
- |
- |
| 15 |
Hb_006120_040 |
0.093673056 |
- |
- |
PREDICTED: prostatic spermine-binding protein [Jatropha curcas] |
| 16 |
Hb_032202_080 |
0.0937320671 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_065525_130 |
0.0937527664 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000847_080 |
0.095259438 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 19 |
Hb_000085_140 |
0.0953602136 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 20 |
Hb_003225_020 |
0.0953873437 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |