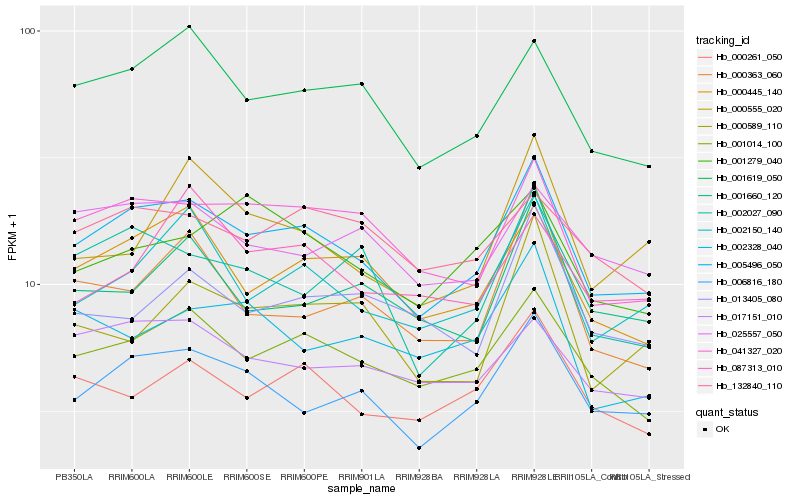
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005496_050 |
0.0 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 2 |
Hb_001014_100 |
0.0695335637 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 3 |
Hb_006816_180 |
0.0717761726 |
- |
- |
- |
| 4 |
Hb_041327_020 |
0.0799137661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000445_140 |
0.0805877212 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 6 |
Hb_001619_050 |
0.0817391886 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002150_140 |
0.0839339271 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 8 |
Hb_000261_050 |
0.0917771553 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 9 |
Hb_002027_090 |
0.0968764912 |
- |
- |
- |
| 10 |
Hb_017151_010 |
0.0972975859 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 11 |
Hb_025557_050 |
0.100173778 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 12 |
Hb_001660_120 |
0.1026161719 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_132840_110 |
0.1031642642 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_001279_040 |
0.1075217508 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 15 |
Hb_000363_060 |
0.1081072788 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000555_020 |
0.1094894126 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_013405_080 |
0.1095917378 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_000589_110 |
0.1106786041 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002328_040 |
0.1124168566 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 20 |
Hb_087313_010 |
0.1165322776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |