| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
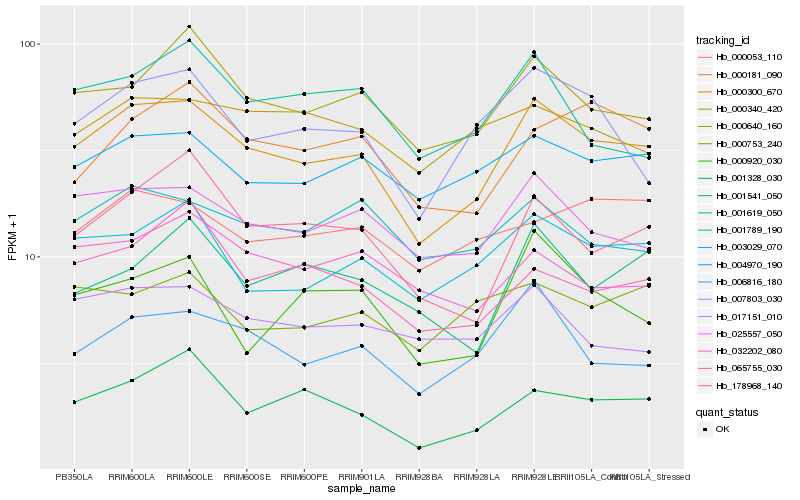
Hb_000300_670 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 2 |
Hb_025557_050 |
0.0920761052 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 3 |
Hb_000640_160 |
0.0930338386 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 4 |
Hb_003029_070 |
0.0990817823 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_004970_190 |
0.0998795552 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_006816_180 |
0.1000898246 |
- |
- |
- |
| 7 |
Hb_007803_030 |
0.101992925 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 8 |
Hb_178968_140 |
0.1020279081 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_001328_030 |
0.1082763456 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 10 |
Hb_001789_190 |
0.1104028111 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 11 |
Hb_000181_090 |
0.111270842 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 12 |
Hb_065755_030 |
0.1142106505 |
- |
- |
- |
| 13 |
Hb_001541_050 |
0.1149000987 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 14 |
Hb_000920_030 |
0.1150920444 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_017151_010 |
0.1160910719 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 16 |
Hb_000340_420 |
0.1168546273 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_032202_080 |
0.1174134257 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000053_110 |
0.1180660636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001619_050 |
0.1181383257 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000753_240 |
0.1185002876 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |