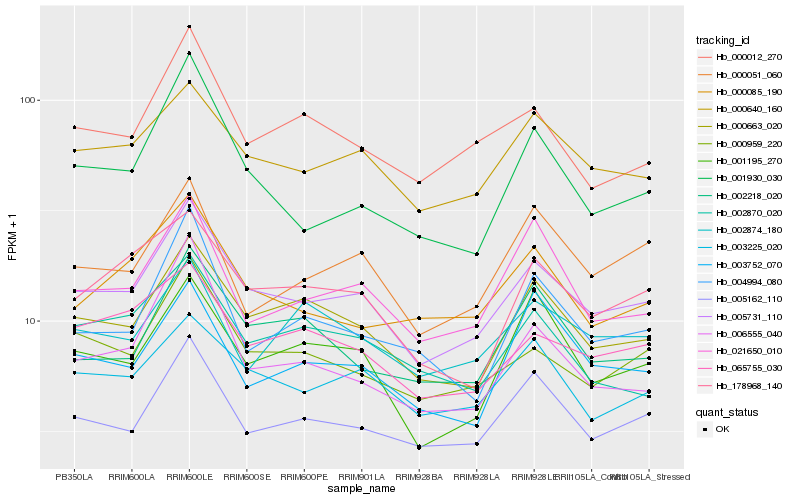
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005731_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 2 |
Hb_000640_160 |
0.0628729823 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 3 |
Hb_000663_020 |
0.064010511 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_000959_220 |
0.0720129326 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_005162_110 |
0.0741805389 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 6 |
Hb_003225_020 |
0.0753481733 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 7 |
Hb_065755_030 |
0.0792856499 |
- |
- |
- |
| 8 |
Hb_000012_270 |
0.0851159074 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 9 |
Hb_178968_140 |
0.0867882289 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_021650_010 |
0.0909686431 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 11 |
Hb_001195_270 |
0.0918489079 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000085_190 |
0.0926487618 |
- |
- |
FtsZ1 protein [Manihot esculenta] |
| 13 |
Hb_002218_020 |
0.0934994216 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 14 |
Hb_001930_030 |
0.0950754571 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000051_060 |
0.0955913973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002870_020 |
0.0975969549 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003752_070 |
0.1003090736 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002874_180 |
0.1005272516 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_004994_080 |
0.1016890345 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_006555_040 |
0.10186181 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |