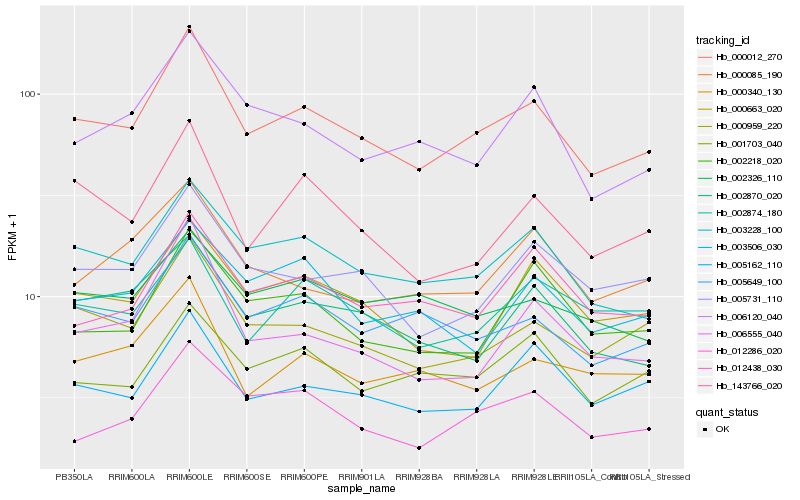
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_270 |
0.0 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 2 |
Hb_003228_100 |
0.0707686314 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 3 |
Hb_000959_220 |
0.0741615565 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_005649_100 |
0.0818996361 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 5 |
Hb_005731_110 |
0.0851159074 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 6 |
Hb_005162_110 |
0.0885717093 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 7 |
Hb_143766_020 |
0.0909833713 |
- |
- |
putative phospholipase A2 [Gossypium arboreum] |
| 8 |
Hb_002870_020 |
0.0913696764 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000663_020 |
0.0925402601 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_006120_040 |
0.0949415101 |
- |
- |
PREDICTED: prostatic spermine-binding protein [Jatropha curcas] |
| 11 |
Hb_012286_020 |
0.0961628579 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 12 |
Hb_001703_040 |
0.0963851803 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000340_130 |
0.0968702758 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 14 |
Hb_012438_030 |
0.0994628264 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 15 |
Hb_002874_180 |
0.0995696893 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002218_020 |
0.1002051665 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 17 |
Hb_006555_040 |
0.1019170007 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 18 |
Hb_002326_110 |
0.1023673919 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 19 |
Hb_000085_190 |
0.1023959922 |
- |
- |
FtsZ1 protein [Manihot esculenta] |
| 20 |
Hb_003506_030 |
0.1041564003 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |