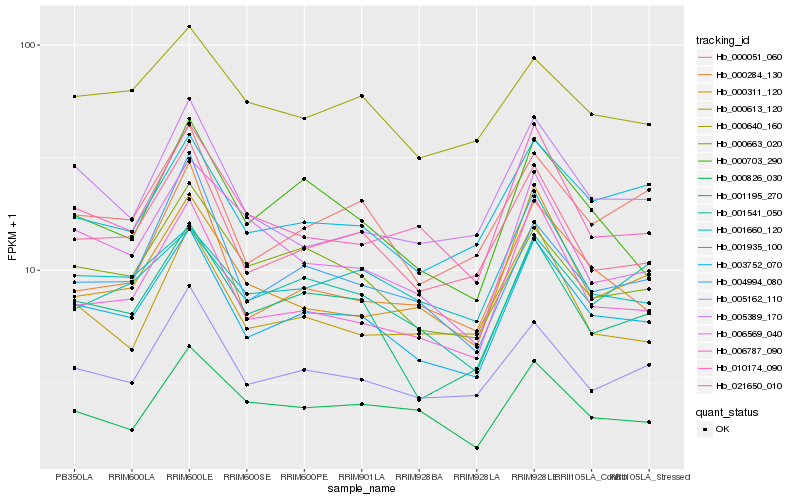
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003752_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 2 |
Hb_021650_010 |
0.0636146832 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 3 |
Hb_001195_270 |
0.0697165281 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006787_090 |
0.0730566657 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 5 |
Hb_005162_110 |
0.0759709477 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 6 |
Hb_000051_060 |
0.0799928377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010174_090 |
0.0818435035 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001935_100 |
0.0827581141 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 9 |
Hb_000703_290 |
0.0863996495 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 10 |
Hb_000663_020 |
0.0864866235 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_005389_170 |
0.0892834361 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 12 |
Hb_000284_130 |
0.0899032169 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 13 |
Hb_000311_120 |
0.0917065971 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 14 |
Hb_001660_120 |
0.0921520131 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000826_030 |
0.093027175 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 16 |
Hb_000640_160 |
0.0942593868 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 17 |
Hb_000613_120 |
0.0954132333 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006569_040 |
0.0970135439 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 19 |
Hb_004994_080 |
0.0971785119 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001541_050 |
0.0979609342 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |