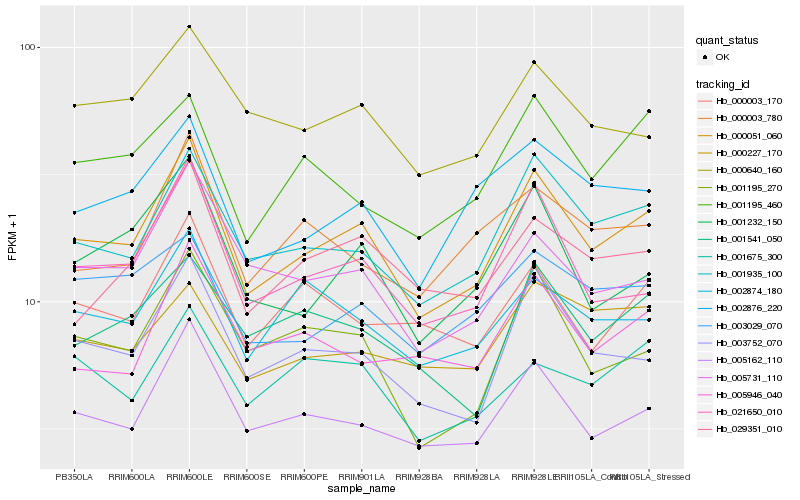
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000051_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001935_100 |
0.067841861 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 3 |
Hb_005162_110 |
0.0742046269 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 4 |
Hb_003752_070 |
0.0799928377 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 5 |
Hb_021650_010 |
0.0806011962 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 6 |
Hb_002876_220 |
0.0914610298 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 7 |
Hb_001232_150 |
0.0927082369 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 8 |
Hb_001195_270 |
0.0953054509 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005731_110 |
0.0955913973 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 10 |
Hb_029351_010 |
0.0956772424 |
- |
- |
PREDICTED: HD domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002874_180 |
0.0970021434 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001195_460 |
0.0972732755 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000640_160 |
0.0989841011 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 14 |
Hb_003029_070 |
0.0993156506 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_001541_050 |
0.1011416556 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 16 |
Hb_000003_170 |
0.1019160767 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 17 |
Hb_000003_780 |
0.1023586605 |
- |
- |
hexokinase [Manihot esculenta] |
| 18 |
Hb_005946_040 |
0.103132502 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001675_300 |
0.1040987072 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000227_170 |
0.1049746545 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |