| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
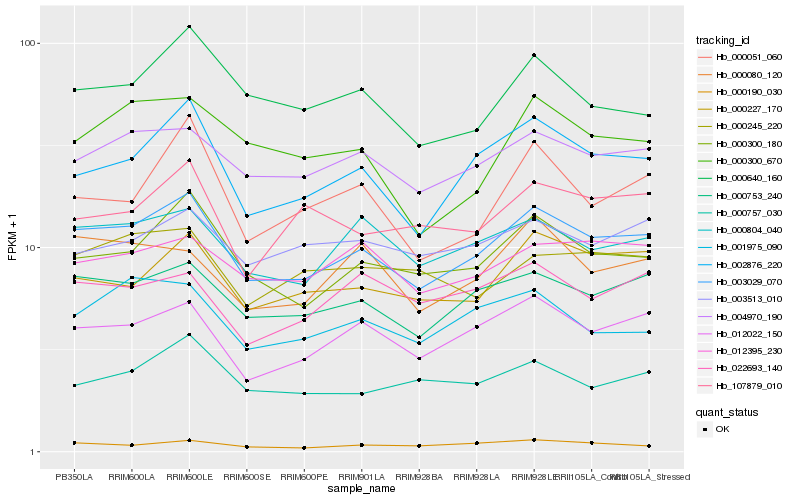
Hb_003029_070 |
0.0 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000753_240 |
0.0595482306 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 3 |
Hb_002876_220 |
0.0681190891 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 4 |
Hb_000300_180 |
0.06943093 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 5 |
Hb_012022_150 |
0.070916232 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 6 |
Hb_004970_190 |
0.0729673018 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000804_040 |
0.0773714896 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000227_170 |
0.0809921243 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_000190_030 |
0.0901128717 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 10 |
Hb_000640_160 |
0.0906186724 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 11 |
Hb_012395_230 |
0.0907522521 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 12 |
Hb_001975_090 |
0.0923356006 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 13 |
Hb_000080_120 |
0.0933618359 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 14 |
Hb_000757_030 |
0.0943863691 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 15 |
Hb_000245_220 |
0.0973946786 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 16 |
Hb_003513_010 |
0.0974858235 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 17 |
Hb_022693_140 |
0.0987830973 |
- |
- |
PREDICTED: uncharacterized protein LOC105632654 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000300_670 |
0.0990817823 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000051_060 |
0.0993156506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_107879_010 |
0.0995158153 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |