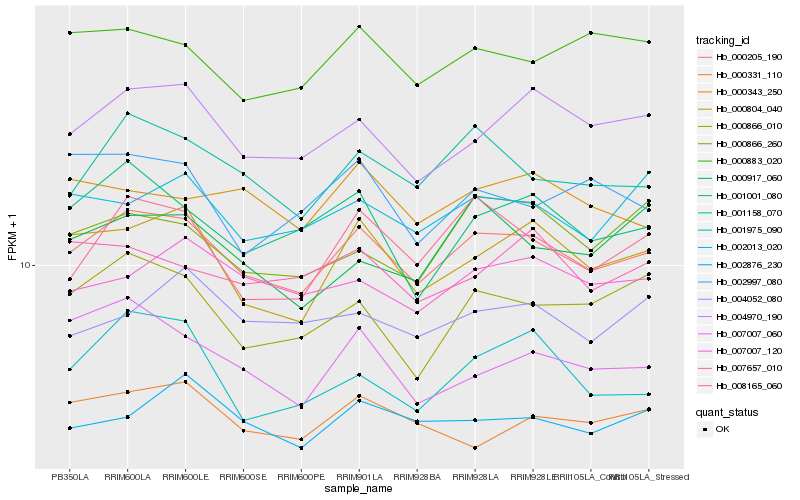
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000205_190 |
0.0 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_001158_070 |
0.055679294 |
- |
- |
Protein DEK, putative [Ricinus communis] |
| 3 |
Hb_004970_190 |
0.0560535657 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000804_040 |
0.0584117651 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_008165_060 |
0.0616399175 |
- |
- |
PREDICTED: uncharacterized protein LOC105635541 [Jatropha curcas] |
| 6 |
Hb_001001_080 |
0.0639959088 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 7 |
Hb_007007_120 |
0.0667485224 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000883_020 |
0.0681037181 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 9 |
Hb_002013_020 |
0.0683394685 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 10 |
Hb_002876_230 |
0.0701625031 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 11 |
Hb_000866_010 |
0.0703180808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007657_010 |
0.0724851244 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000866_260 |
0.0737388305 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000331_110 |
0.0743279425 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 15 |
Hb_000343_250 |
0.0751833528 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 16 |
Hb_004052_080 |
0.0754592273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007007_060 |
0.075930058 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001975_090 |
0.0776609213 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 19 |
Hb_000917_060 |
0.0780001803 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 20 |
Hb_002997_080 |
0.078916925 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |