| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
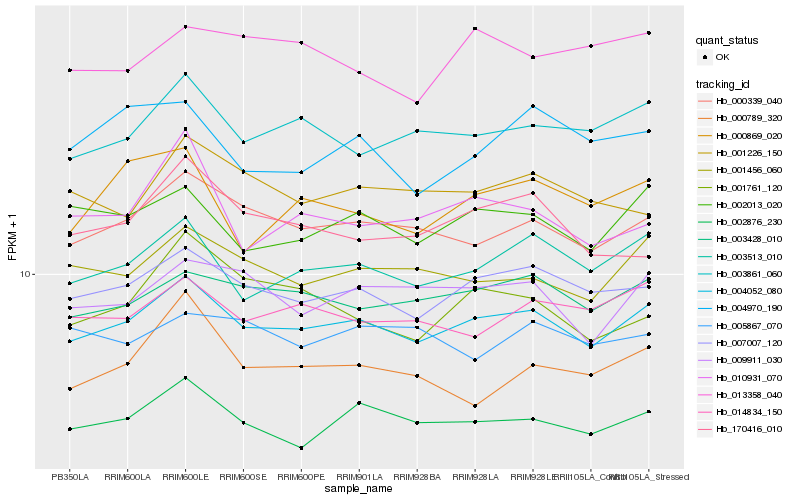
Hb_004052_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007007_120 |
0.0394214899 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000339_040 |
0.0448139514 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 4 |
Hb_003513_010 |
0.0461080948 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 5 |
Hb_002876_230 |
0.0475576093 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 6 |
Hb_003428_010 |
0.0477353555 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 7 |
Hb_003861_060 |
0.053352333 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 8 |
Hb_001456_060 |
0.053894437 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 9 |
Hb_002013_020 |
0.0548498804 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 10 |
Hb_009911_030 |
0.057289458 |
transcription factor |
TF Family: FAR1 |
far-red impaired response 1 -like protein [Gossypium arboreum] |
| 11 |
Hb_014834_150 |
0.0581252043 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_001226_150 |
0.0695217736 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 13 |
Hb_170416_010 |
0.0696126052 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 14 |
Hb_000869_020 |
0.0699307147 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 15 |
Hb_013358_040 |
0.0719059304 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 16 |
Hb_000789_320 |
0.0728521323 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_010931_070 |
0.0731947183 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001761_120 |
0.0733166536 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 19 |
Hb_004970_190 |
0.0734116389 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_005867_070 |
0.0739957573 |
- |
- |
DNA binding protein, putative [Ricinus communis] |