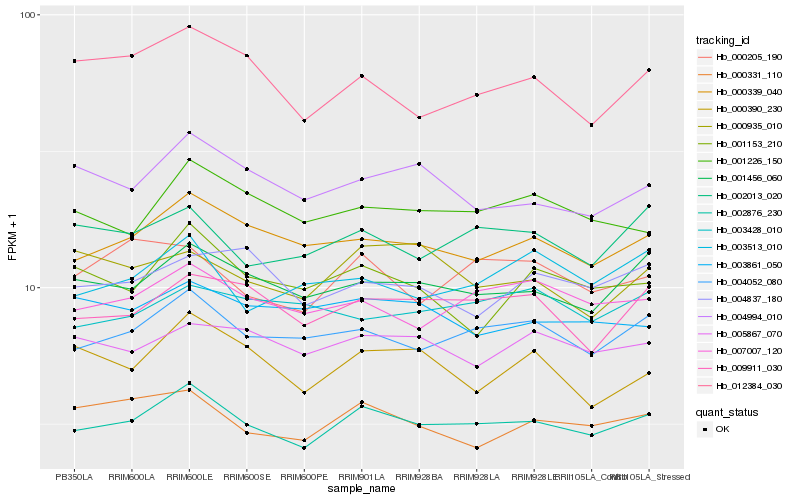
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 2 |
Hb_004052_080 |
0.0475576093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000339_040 |
0.052212756 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 4 |
Hb_001456_060 |
0.0539632309 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 5 |
Hb_007007_120 |
0.0585803299 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009911_030 |
0.0606667853 |
transcription factor |
TF Family: FAR1 |
far-red impaired response 1 -like protein [Gossypium arboreum] |
| 7 |
Hb_000331_110 |
0.064965546 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 8 |
Hb_002013_020 |
0.0651530545 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 9 |
Hb_001226_150 |
0.0660973933 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 10 |
Hb_005867_070 |
0.0676095124 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_004994_010 |
0.0688094862 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 12 |
Hb_000205_190 |
0.0701625031 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_003513_010 |
0.07109333 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 14 |
Hb_012384_030 |
0.0718519102 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 15 |
Hb_003861_050 |
0.0743632635 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 16 |
Hb_004837_180 |
0.0753206901 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003428_010 |
0.0761721059 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 18 |
Hb_001153_210 |
0.0767225409 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000390_230 |
0.0770834285 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 20 |
Hb_000935_010 |
0.0770905056 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |