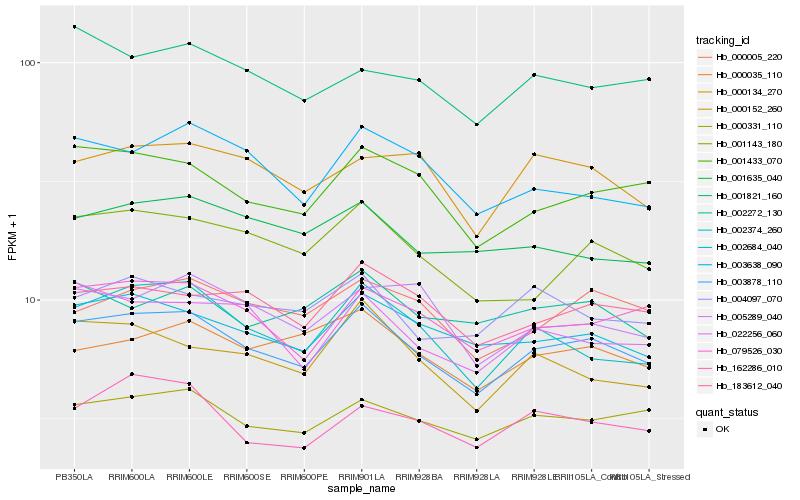
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003878_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 2 |
Hb_002684_040 |
0.060144028 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001433_070 |
0.0616154362 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 4 |
Hb_000331_110 |
0.0632787288 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 5 |
Hb_079526_030 |
0.0632805904 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 6 |
Hb_002374_260 |
0.0641994223 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 7 |
Hb_001143_180 |
0.0643832021 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 8 |
Hb_183612_040 |
0.0654276316 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 9 |
Hb_022256_060 |
0.068562853 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 10 |
Hb_000152_260 |
0.07098717 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 11 |
Hb_000005_220 |
0.0712932034 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 12 |
Hb_003638_090 |
0.0729638093 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 13 |
Hb_000035_110 |
0.0746645263 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 14 |
Hb_004097_070 |
0.0755171954 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 15 |
Hb_001635_040 |
0.0762373792 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_005289_040 |
0.0776447281 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 17 |
Hb_000134_270 |
0.0777256593 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 18 |
Hb_162286_010 |
0.0778302592 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 19 |
Hb_002272_130 |
0.0784686676 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 20 |
Hb_001821_160 |
0.0792996725 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |