| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
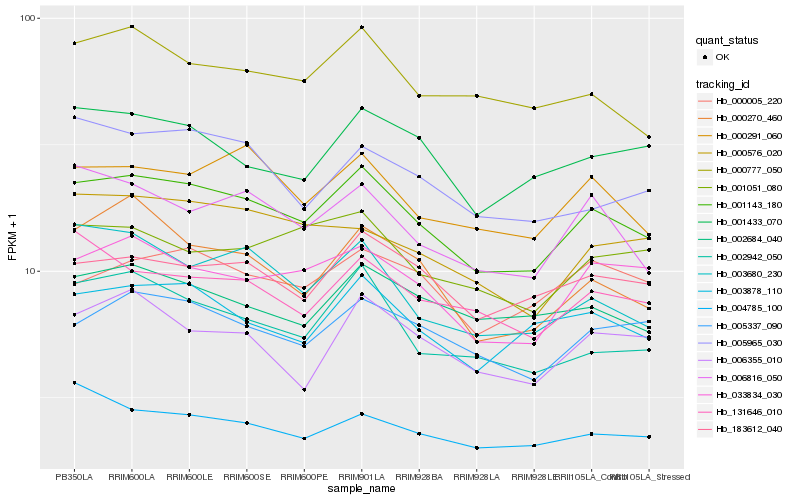
Hb_001143_180 |
0.0 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 2 |
Hb_003878_110 |
0.0643832021 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 3 |
Hb_000291_060 |
0.0645535472 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_033834_030 |
0.0652645716 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_006816_050 |
0.066837971 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 6 |
Hb_183612_040 |
0.0672857725 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 7 |
Hb_005337_090 |
0.0678503253 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 8 |
Hb_002942_050 |
0.0708750207 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000777_050 |
0.0715670578 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 10 |
Hb_001433_070 |
0.0724780199 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 11 |
Hb_002684_040 |
0.0736054536 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 12 |
Hb_131646_010 |
0.0742514579 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000270_460 |
0.0748343711 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 14 |
Hb_000005_220 |
0.0754305455 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 15 |
Hb_001051_080 |
0.075452728 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 16 |
Hb_003680_230 |
0.0756389255 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 17 |
Hb_000576_020 |
0.0766303564 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 18 |
Hb_006355_010 |
0.0774338293 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 19 |
Hb_005965_030 |
0.0784151931 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 20 |
Hb_004785_100 |
0.0789318266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |