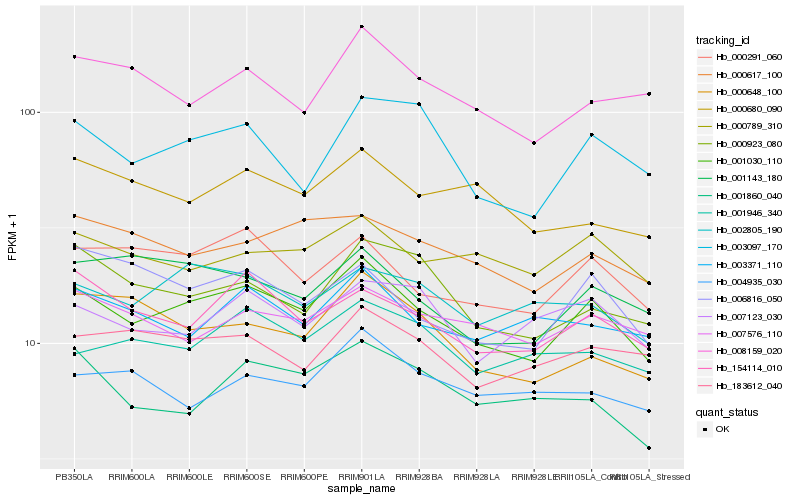
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001030_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000291_060 |
0.0752076389 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_154114_010 |
0.0805904641 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 4 |
Hb_000617_100 |
0.0850909579 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 5 |
Hb_000789_310 |
0.0853545157 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PRT1 [Jatropha curcas] |
| 6 |
Hb_002805_190 |
0.0859910047 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 7 |
Hb_001143_180 |
0.0877494724 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 8 |
Hb_003097_170 |
0.0877650499 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_007576_110 |
0.0891372998 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 10 |
Hb_183612_040 |
0.0896015045 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 11 |
Hb_001946_340 |
0.0905619495 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000923_080 |
0.0906414799 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 13 |
Hb_003371_110 |
0.0910982694 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 14 |
Hb_000680_090 |
0.0914457495 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 15 |
Hb_006816_050 |
0.0915767972 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 16 |
Hb_001860_040 |
0.0916916747 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X4 [Jatropha curcas] |
| 17 |
Hb_008159_020 |
0.0917571054 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 18 |
Hb_007123_030 |
0.0922804935 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000648_100 |
0.0943966908 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 20 |
Hb_004935_030 |
0.0959415548 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |