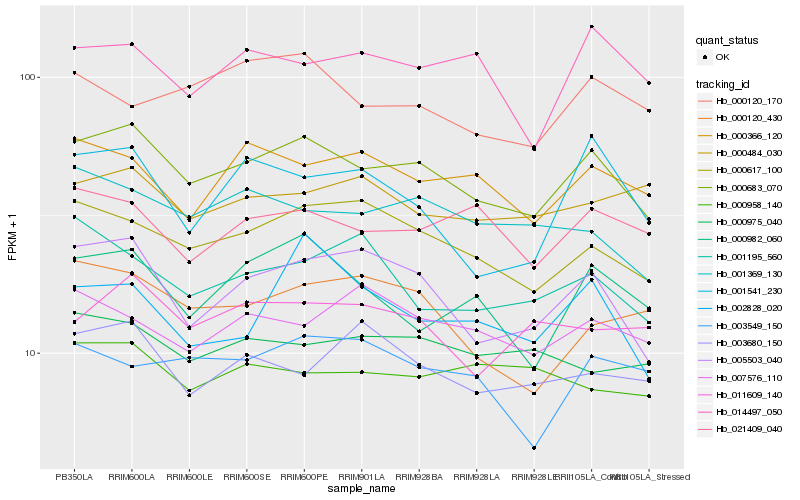
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000683_070 |
0.0 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_005503_040 |
0.0659917693 |
- |
- |
PREDICTED: AMP deaminase [Jatropha curcas] |
| 3 |
Hb_000617_100 |
0.0661910746 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 4 |
Hb_021409_040 |
0.0809173868 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 5 |
Hb_000366_120 |
0.0809219798 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 6 |
Hb_001369_130 |
0.0810577328 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000982_060 |
0.081263896 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 8 |
Hb_014497_050 |
0.0854067917 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_08021 [Jatropha curcas] |
| 9 |
Hb_011609_140 |
0.0857582216 |
- |
- |
hypothetical protein JCGZ_12974 [Jatropha curcas] |
| 10 |
Hb_000120_170 |
0.0872943393 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 11 |
Hb_000975_040 |
0.088341625 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 12 |
Hb_000120_430 |
0.0898835388 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 13 |
Hb_007576_110 |
0.0908248056 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 14 |
Hb_001541_230 |
0.0911028222 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_003680_150 |
0.0913964154 |
- |
- |
srpk, putative [Ricinus communis] |
| 16 |
Hb_003549_150 |
0.0915371248 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 17 |
Hb_000484_030 |
0.0916155223 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 18 |
Hb_001195_560 |
0.0921035575 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000958_140 |
0.09226745 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |
| 20 |
Hb_002828_020 |
0.0924274356 |
- |
- |
protein binding protein, putative [Ricinus communis] |