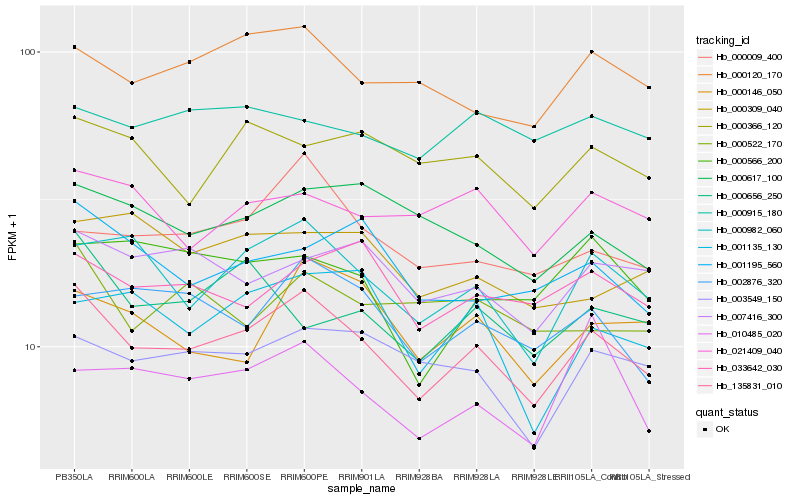
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135831_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_13374 [Jatropha curcas] |
| 2 |
Hb_000982_060 |
0.073694732 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 3 |
Hb_000120_170 |
0.0854660082 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 4 |
Hb_003549_150 |
0.0890563712 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 5 |
Hb_002876_320 |
0.0891444781 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 6 |
Hb_007416_300 |
0.0908500646 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 7 |
Hb_000009_400 |
0.0915964736 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 8 |
Hb_033642_030 |
0.0925817967 |
- |
- |
PREDICTED: G-protein coupled receptor 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001195_560 |
0.0937702787 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000617_100 |
0.0938625086 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 11 |
Hb_000146_050 |
0.0942931744 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |
| 12 |
Hb_021409_040 |
0.0953296714 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 13 |
Hb_000366_120 |
0.0968209452 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 14 |
Hb_001135_130 |
0.0994435331 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |
| 15 |
Hb_000522_170 |
0.0995155103 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 16 |
Hb_000309_040 |
0.1010784008 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 17 |
Hb_010485_020 |
0.1013621856 |
- |
- |
PREDICTED: thaumatin-like protein 1b isoform X1 [Jatropha curcas] |
| 18 |
Hb_000566_200 |
0.1016466159 |
- |
- |
PREDICTED: uncharacterized protein LOC105644834 [Jatropha curcas] |
| 19 |
Hb_000656_250 |
0.1017214881 |
- |
- |
hypothetical protein JCGZ_24680 [Jatropha curcas] |
| 20 |
Hb_000915_180 |
0.1021441124 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Pyrus x bretschneideri] |