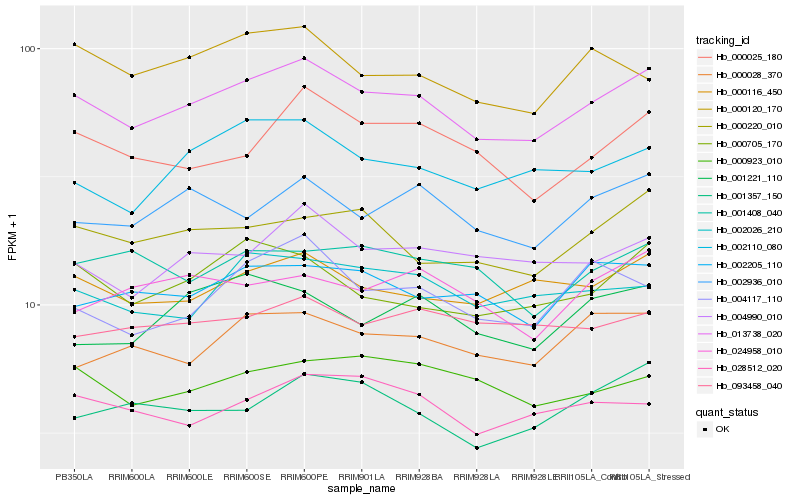
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013738_020 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 2 |
Hb_000116_450 |
0.0615040018 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_000220_010 |
0.065369583 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |
| 4 |
Hb_000705_170 |
0.0667102498 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 5 |
Hb_002936_010 |
0.0672892402 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |
| 6 |
Hb_004990_010 |
0.0673856857 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_002110_080 |
0.0678098071 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 8 |
Hb_000025_180 |
0.0683488665 |
- |
- |
hypothetical protein CISIN_1g024241mg [Citrus sinensis] |
| 9 |
Hb_002026_210 |
0.0704046282 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 10 |
Hb_002205_110 |
0.0712026079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_028512_020 |
0.0713532617 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000923_010 |
0.0729498298 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000028_370 |
0.0729684104 |
- |
- |
Guanine nucleotide-binding protein alpha-2 subunit [Glycine soja] |
| 14 |
Hb_001357_150 |
0.0741094258 |
- |
- |
hypothetical protein POPTR_0011s08930g [Populus trichocarpa] |
| 15 |
Hb_000120_170 |
0.0743747827 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 16 |
Hb_004117_110 |
0.0766802534 |
- |
- |
hypothetical protein POPTR_0016s14420g [Populus trichocarpa] |
| 17 |
Hb_001408_040 |
0.077029555 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 18 |
Hb_093458_040 |
0.0775715946 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 19 |
Hb_001221_110 |
0.0781492558 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 20 |
Hb_024958_010 |
0.0781687183 |
- |
- |
PREDICTED: LRR repeats and ubiquitin-like domain-containing protein At2g30105 [Jatropha curcas] |