| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
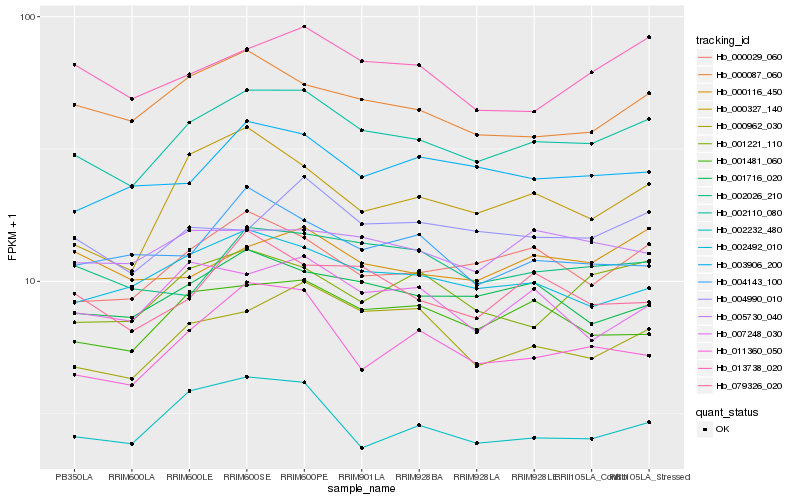
Hb_002110_080 |
0.0 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 2 |
Hb_000029_060 |
0.0601406073 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 3 |
Hb_001481_060 |
0.0624372581 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002492_010 |
0.0649694384 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 5 |
Hb_001716_020 |
0.0651830352 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 6 |
Hb_000087_060 |
0.0656485226 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 7 |
Hb_000962_030 |
0.0660760863 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 8 |
Hb_004990_010 |
0.0676926568 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |
| 9 |
Hb_013738_020 |
0.0678098071 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 10 |
Hb_003906_200 |
0.0693567801 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 11 |
Hb_079326_020 |
0.0702926603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002026_210 |
0.0715178796 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 13 |
Hb_001221_110 |
0.0715305308 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 14 |
Hb_011360_050 |
0.0731294715 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005730_040 |
0.073163564 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 16 |
Hb_000327_140 |
0.0737923409 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007248_030 |
0.0740754297 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 18 |
Hb_000116_450 |
0.0755610808 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_002232_480 |
0.0764615612 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 20 |
Hb_004143_100 |
0.0776541554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |