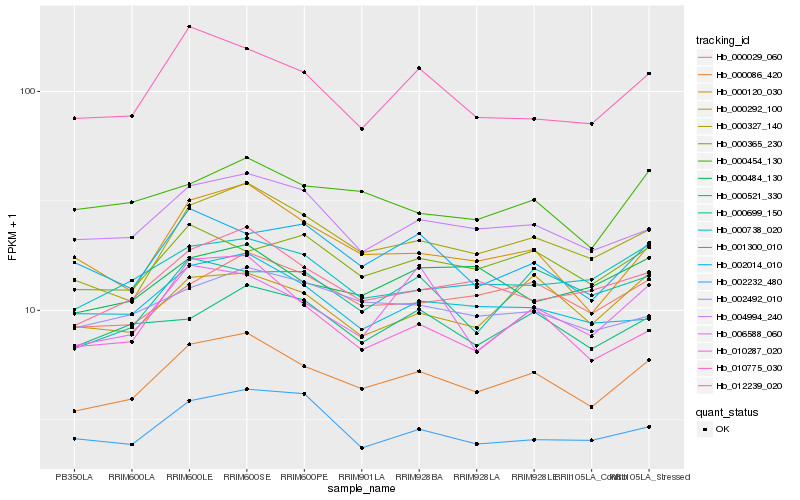
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_420 |
0.0 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 2 |
Hb_000327_140 |
0.0533758194 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000029_060 |
0.061827309 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 4 |
Hb_012239_020 |
0.0636182839 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 5 |
Hb_004994_240 |
0.0656587232 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 6 |
Hb_000292_100 |
0.0710810201 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 7 |
Hb_000738_020 |
0.0727473011 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000120_030 |
0.073004602 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002492_010 |
0.0751752456 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 10 |
Hb_000365_230 |
0.0755072166 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 11 |
Hb_000521_330 |
0.0756374839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010775_030 |
0.0758815995 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 13 |
Hb_001300_010 |
0.076046756 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_000454_130 |
0.0783146759 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006588_060 |
0.0792764548 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 16 |
Hb_010287_020 |
0.0809962324 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 17 |
Hb_002232_480 |
0.08155797 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 18 |
Hb_000484_130 |
0.0818096286 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 19 |
Hb_002014_010 |
0.0821121457 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000699_150 |
0.0827890428 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |