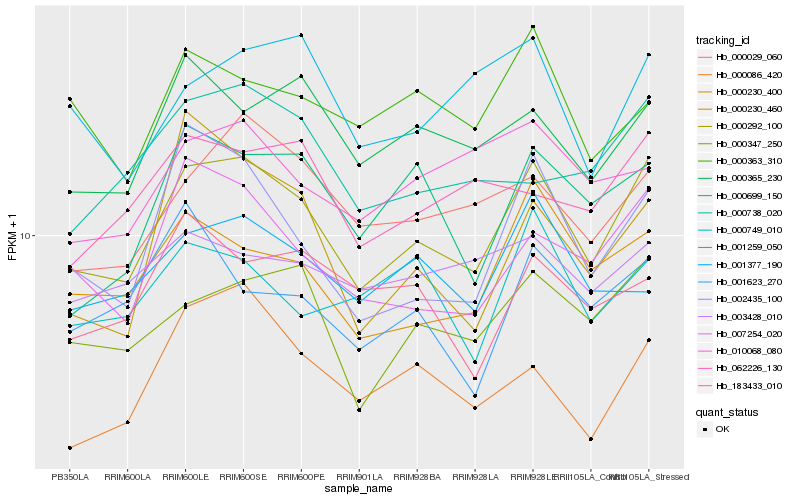
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000292_100 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 2 |
Hb_000347_250 |
0.0621341384 |
- |
- |
PREDICTED: uncharacterized protein LOC105133971 [Populus euphratica] |
| 3 |
Hb_007254_020 |
0.0645512597 |
- |
- |
PREDICTED: uncharacterized protein LOC105629137 [Jatropha curcas] |
| 4 |
Hb_000230_400 |
0.0661973263 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_002435_100 |
0.067638506 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 6 |
Hb_000029_060 |
0.0680224481 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 7 |
Hb_000086_420 |
0.0710810201 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 8 |
Hb_000365_230 |
0.0712593213 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 9 |
Hb_000699_150 |
0.0760201135 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 10 |
Hb_062226_130 |
0.0777434615 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 11 |
Hb_000738_020 |
0.0779007442 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_001377_190 |
0.0795114611 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001259_050 |
0.0801241527 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 14 |
Hb_010068_080 |
0.0805757275 |
- |
- |
PREDICTED: RNA-binding protein rsd1-like [Jatropha curcas] |
| 15 |
Hb_000363_310 |
0.0806589685 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 16 |
Hb_000749_010 |
0.0811090513 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003428_010 |
0.0815397738 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 18 |
Hb_183433_010 |
0.0823727941 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 19 |
Hb_001623_270 |
0.0827158895 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 20 |
Hb_000230_460 |
0.0829295465 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |