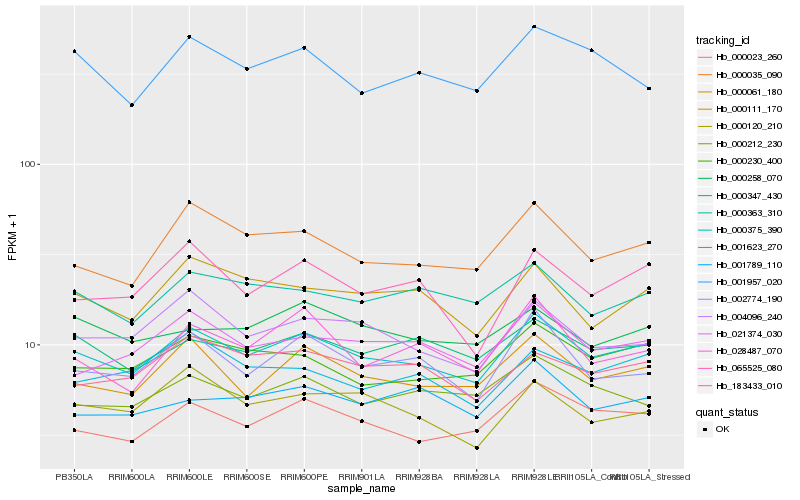
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000375_390 |
0.0 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 2 |
Hb_000023_260 |
0.0566327702 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000061_180 |
0.0578668155 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 4 |
Hb_000035_090 |
0.0645379907 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 5 |
Hb_000230_400 |
0.0660941378 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000347_430 |
0.0698599021 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 7 |
Hb_021374_030 |
0.0707384301 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 8 |
Hb_004096_240 |
0.0714036166 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 9 |
Hb_002774_190 |
0.0720150862 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 10 |
Hb_000363_310 |
0.0721491681 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 11 |
Hb_000111_170 |
0.0756323437 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 12 |
Hb_065525_080 |
0.0768551535 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 13 |
Hb_183433_010 |
0.0769813286 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 14 |
Hb_001957_020 |
0.0771172217 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 15 |
Hb_000212_230 |
0.0777915728 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 16 |
Hb_000258_070 |
0.0790868915 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 17 |
Hb_028487_070 |
0.0804831153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000120_210 |
0.0806874915 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 19 |
Hb_001789_110 |
0.0809490549 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_001623_270 |
0.0811274179 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |