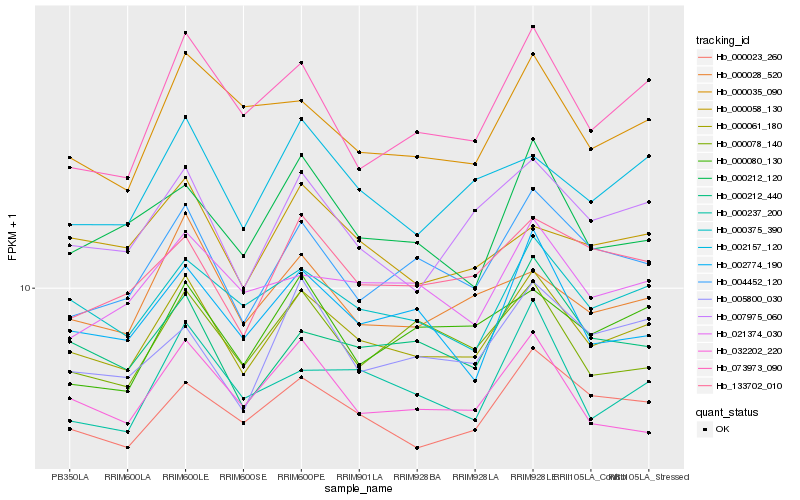
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_180 |
0.0 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 2 |
Hb_000375_390 |
0.0578668155 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 3 |
Hb_000023_260 |
0.0611510774 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007975_060 |
0.0640536101 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004452_120 |
0.0652182838 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 6 |
Hb_133702_010 |
0.0704669263 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_002157_120 |
0.0705849511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000028_520 |
0.0718254204 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 9 |
Hb_000212_440 |
0.0719648967 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_032202_220 |
0.0739457202 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 11 |
Hb_002774_190 |
0.0743509938 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 12 |
Hb_021374_030 |
0.0743782043 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 13 |
Hb_000035_090 |
0.0751081305 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 14 |
Hb_073973_090 |
0.0763506664 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 15 |
Hb_000078_140 |
0.0768204896 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 16 |
Hb_000212_120 |
0.077153681 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 17 |
Hb_000237_200 |
0.0776349468 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 18 |
Hb_005800_030 |
0.0782480321 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 19 |
Hb_000080_130 |
0.0785049626 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 20 |
Hb_000058_130 |
0.0789303198 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |