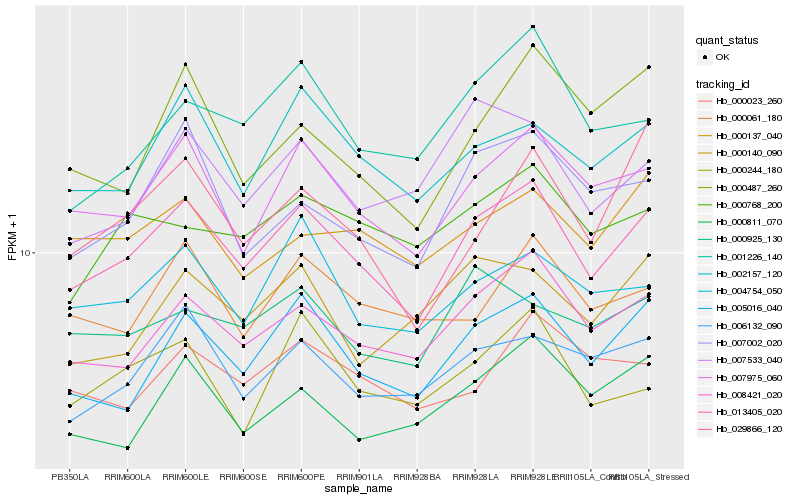
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013405_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_007975_060 |
0.0657468204 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 3 |
Hb_008421_020 |
0.0667244816 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005016_040 |
0.0728392801 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 5 |
Hb_002157_120 |
0.0809427261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006132_090 |
0.085963765 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004754_050 |
0.0891267622 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 8 |
Hb_007002_020 |
0.0935848965 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 9 |
Hb_000061_180 |
0.0941951719 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 10 |
Hb_001226_140 |
0.0943216562 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 11 |
Hb_000137_040 |
0.0965600262 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 12 |
Hb_000768_200 |
0.0969733319 |
- |
- |
hypothetical protein POPTR_0013s06910g [Populus trichocarpa] |
| 13 |
Hb_007533_040 |
0.0977330918 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_000925_130 |
0.0980065938 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000023_260 |
0.0986562762 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000811_070 |
0.0995749181 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 17 |
Hb_000244_180 |
0.1011513207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000140_090 |
0.1014251302 |
- |
- |
PREDICTED: C-terminal binding protein AN [Jatropha curcas] |
| 19 |
Hb_000487_260 |
0.1035368986 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 20 |
Hb_029866_120 |
0.1036118242 |
- |
- |
hypothetical protein CICLE_v10006070mg [Citrus clementina] |