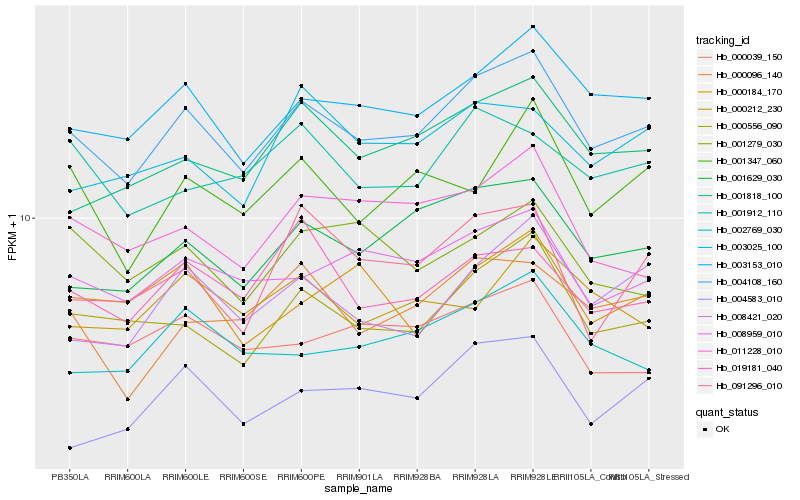
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004108_160 |
0.0 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 2 |
Hb_003153_010 |
0.0517002604 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 3 |
Hb_000556_090 |
0.0590158205 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001629_030 |
0.0610093446 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 5 |
Hb_008959_010 |
0.0655765198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001818_100 |
0.0690550585 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 7 |
Hb_008421_020 |
0.0721784093 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002769_030 |
0.0769840339 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000096_140 |
0.0774377239 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_019181_040 |
0.0784254978 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 11 |
Hb_011228_010 |
0.0786491791 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_001347_060 |
0.0796979846 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 13 |
Hb_001912_110 |
0.0809427153 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_000039_150 |
0.0810053785 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003025_100 |
0.0810802656 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004583_010 |
0.0833096818 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22150, chloroplastic [Jatropha curcas] |
| 17 |
Hb_091296_010 |
0.084073523 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 18 |
Hb_000184_170 |
0.0848136234 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 19 |
Hb_000212_230 |
0.0852207255 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 20 |
Hb_001279_030 |
0.0853407742 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |