| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
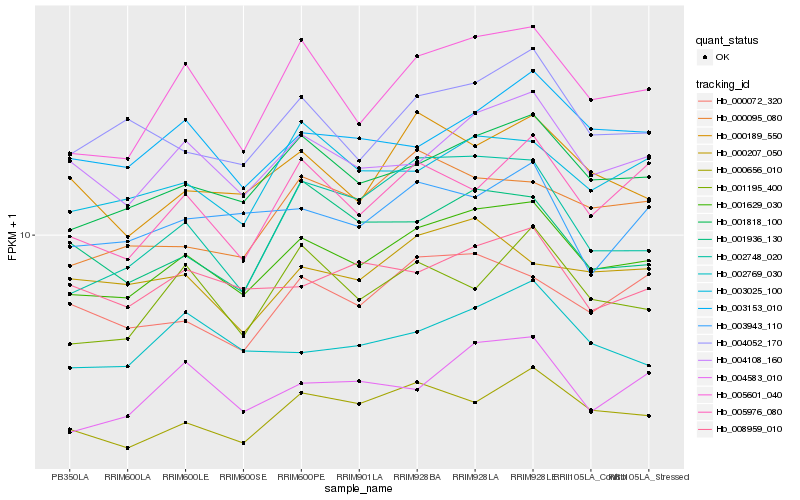
Hb_001629_030 |
0.0 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 2 |
Hb_001818_100 |
0.0461929101 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 3 |
Hb_005601_040 |
0.0593766486 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 4 |
Hb_004108_160 |
0.0610093446 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 5 |
Hb_000656_010 |
0.0690068882 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004052_170 |
0.0709783456 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_003153_010 |
0.0731506354 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 8 |
Hb_003025_100 |
0.0735530729 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002769_030 |
0.0768498682 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_008959_010 |
0.0778079444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000189_550 |
0.0782690508 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 12 |
Hb_000072_320 |
0.0783816403 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 13 |
Hb_004583_010 |
0.0785715696 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22150, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005976_080 |
0.0795083017 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 15 |
Hb_003943_110 |
0.0805339333 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 16 |
Hb_000095_080 |
0.0833454878 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 17 |
Hb_002748_020 |
0.0833572894 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 18 |
Hb_001195_400 |
0.0842021707 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 19 |
Hb_001936_130 |
0.0848349002 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 20 |
Hb_000207_050 |
0.0849714986 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |