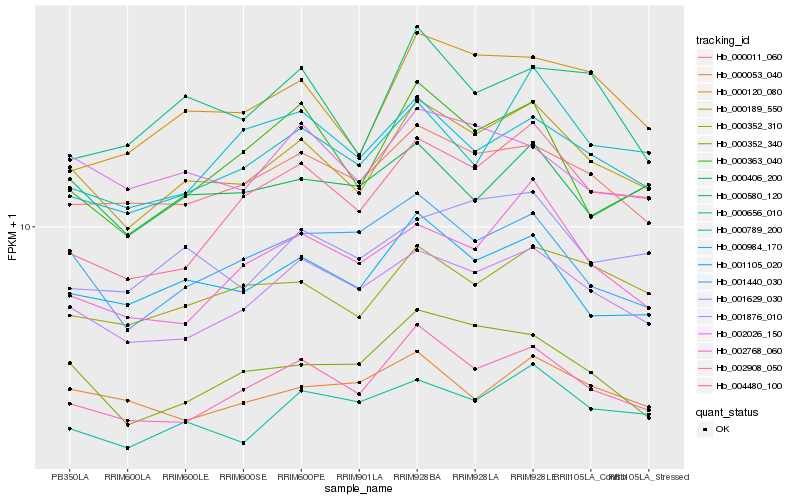
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_550 |
0.0 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 2 |
Hb_001876_010 |
0.0585651961 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002908_050 |
0.0599598654 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 4 |
Hb_001105_020 |
0.0659671768 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 5 |
Hb_000011_060 |
0.066931459 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000352_310 |
0.0689848634 |
- |
- |
hypothetical protein CISIN_1g0017891mg, partial [Citrus sinensis] |
| 7 |
Hb_000656_010 |
0.0720715639 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000789_200 |
0.0775552866 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 9 |
Hb_000120_080 |
0.0777509867 |
- |
- |
PREDICTED: probable pyridoxal 5'-phosphate synthase subunit PDX1 [Jatropha curcas] |
| 10 |
Hb_001629_030 |
0.0782690508 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 11 |
Hb_002026_150 |
0.0785975793 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair helicase XPB1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001440_030 |
0.0822549286 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 13 |
Hb_000352_340 |
0.0826523692 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 14 |
Hb_000406_200 |
0.0827538106 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 15 |
Hb_000363_040 |
0.0840467645 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 16 |
Hb_004480_100 |
0.0844700071 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 17 |
Hb_000984_170 |
0.0847873416 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 18 |
Hb_002768_060 |
0.0860302025 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000580_120 |
0.0862893773 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000053_040 |
0.0876560395 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |