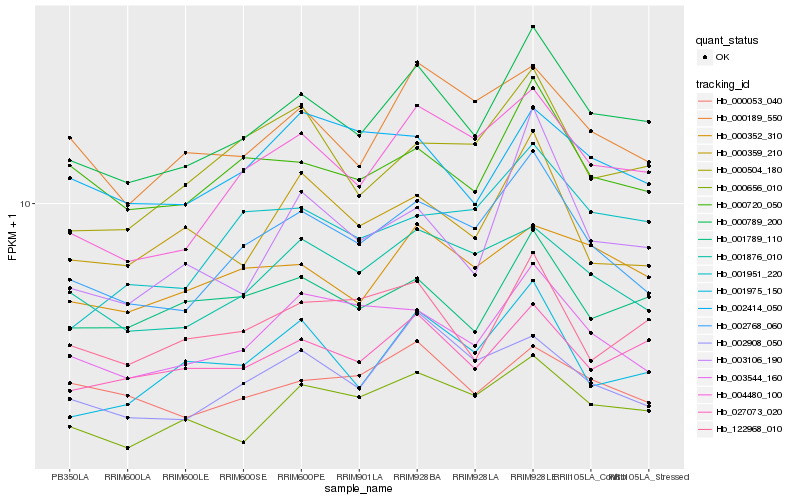
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000789_200 |
0.0 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 2 |
Hb_027073_020 |
0.0603336207 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 3 |
Hb_002768_060 |
0.066243322 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000656_010 |
0.067815522 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004480_100 |
0.0680501973 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 6 |
Hb_000720_050 |
0.0713282024 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001789_110 |
0.0717611953 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_000352_310 |
0.0727738545 |
- |
- |
hypothetical protein CISIN_1g0017891mg, partial [Citrus sinensis] |
| 9 |
Hb_000189_550 |
0.0775552866 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 10 |
Hb_122968_010 |
0.0775951253 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 11 |
Hb_003544_160 |
0.0781584933 |
- |
- |
- |
| 12 |
Hb_001876_010 |
0.0782314937 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001975_150 |
0.0815156035 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 14 |
Hb_000053_040 |
0.0843043909 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 15 |
Hb_000504_180 |
0.0848450009 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 16 |
Hb_002908_050 |
0.0853533732 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 17 |
Hb_000359_210 |
0.0864324065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003106_190 |
0.0871418902 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 19 |
Hb_001951_220 |
0.0883005561 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 20 |
Hb_002414_050 |
0.0883693092 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |