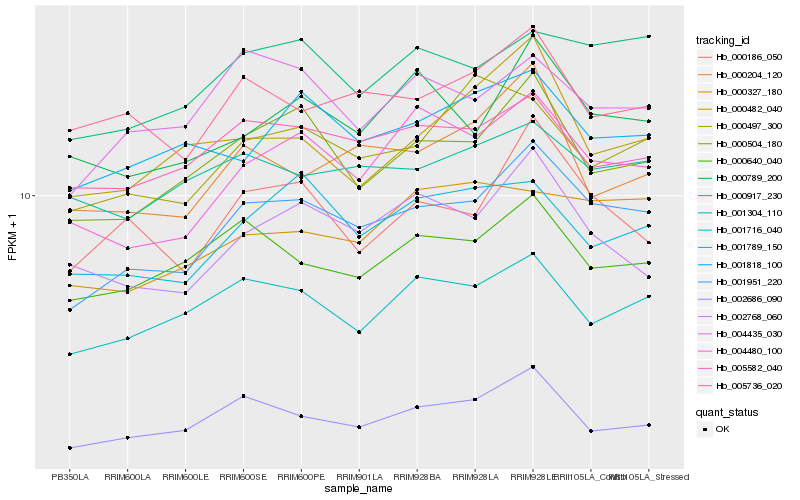
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_220 |
0.0 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 2 |
Hb_000504_180 |
0.0723171033 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 3 |
Hb_004480_100 |
0.0781834336 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 4 |
Hb_000917_230 |
0.0794018602 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 5 |
Hb_002686_090 |
0.0806822416 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 6 |
Hb_001716_040 |
0.0843268147 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 7 |
Hb_000186_050 |
0.0845183454 |
- |
- |
PREDICTED: uncharacterized protein LOC105649281 [Jatropha curcas] |
| 8 |
Hb_005582_040 |
0.0871328136 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 9 |
Hb_004435_030 |
0.0872951644 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 10 |
Hb_000789_200 |
0.0883005561 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_000640_040 |
0.0898354241 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001818_100 |
0.0907334179 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 13 |
Hb_002768_060 |
0.0916280038 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005736_020 |
0.0921118505 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 15 |
Hb_001789_150 |
0.0928241278 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000204_120 |
0.0939084426 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 17 |
Hb_001304_110 |
0.0959199121 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 18 |
Hb_000327_180 |
0.0962653859 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000497_300 |
0.0965923141 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 20 |
Hb_000482_040 |
0.0980934657 |
- |
- |
Protein YME1, putative [Ricinus communis] |