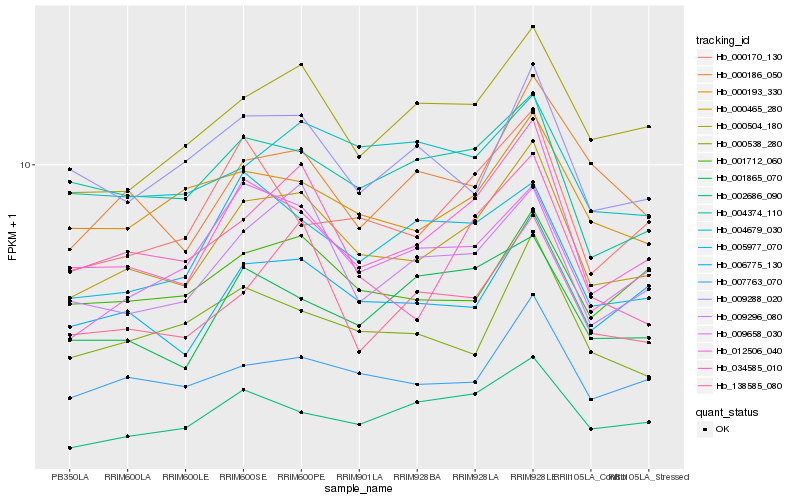
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_280 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 2 |
Hb_012506_040 |
0.0569067962 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 3 |
Hb_006775_130 |
0.0706262764 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 4 |
Hb_007763_070 |
0.0747265622 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000504_180 |
0.076638395 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_009296_080 |
0.0767036978 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002686_090 |
0.0826529871 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 8 |
Hb_009658_030 |
0.082684084 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 9 |
Hb_000193_330 |
0.08423588 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 10 |
Hb_001712_060 |
0.0864502897 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 11 |
Hb_004374_110 |
0.0867373445 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 12 |
Hb_138585_080 |
0.0889203594 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 13 |
Hb_000170_130 |
0.0911846423 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 14 |
Hb_000186_050 |
0.0915459765 |
- |
- |
PREDICTED: uncharacterized protein LOC105649281 [Jatropha curcas] |
| 15 |
Hb_001865_070 |
0.0923780041 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005977_070 |
0.0929452609 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000538_280 |
0.0929562331 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 18 |
Hb_009288_020 |
0.0947025224 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 19 |
Hb_004679_030 |
0.0957009314 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 20 |
Hb_034585_010 |
0.0963259966 |
- |
- |
PREDICTED: uncharacterized protein LOC105639637 [Jatropha curcas] |