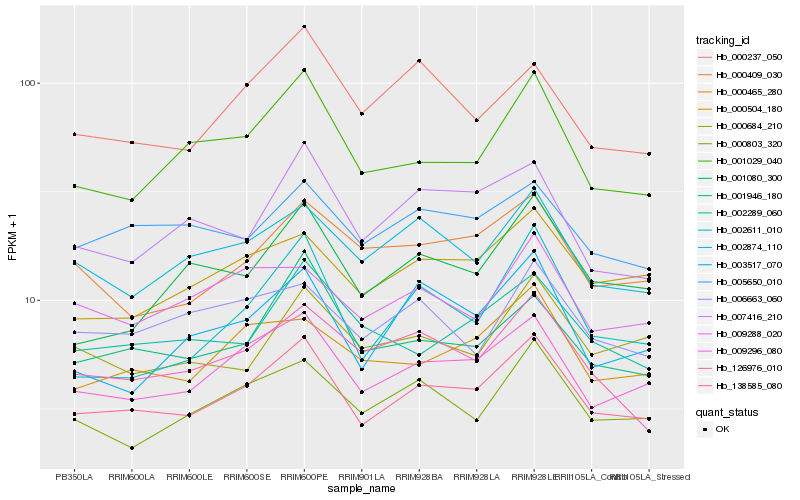
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138585_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 2 |
Hb_009296_080 |
0.0555899329 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001029_040 |
0.0788356119 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 4 |
Hb_000504_180 |
0.0854121579 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 5 |
Hb_001946_180 |
0.0870001231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002874_110 |
0.0871359365 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 7 |
Hb_000465_280 |
0.0889203594 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 8 |
Hb_000803_320 |
0.0891480708 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 9 |
Hb_005650_010 |
0.093795387 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_007416_210 |
0.0938242519 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 11 |
Hb_126976_010 |
0.0945470091 |
- |
- |
- |
| 12 |
Hb_001080_300 |
0.0951233357 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 13 |
Hb_002289_060 |
0.0956250389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000409_030 |
0.0959104343 |
- |
- |
PREDICTED: uncharacterized protein LOC105642693 [Jatropha curcas] |
| 15 |
Hb_003517_070 |
0.0966083343 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 16 |
Hb_002611_010 |
0.099289463 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_009288_020 |
0.0995015741 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 18 |
Hb_000237_050 |
0.0996794648 |
- |
- |
CP2 [Hevea brasiliensis] |
| 19 |
Hb_000684_210 |
0.1005011761 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 20 |
Hb_006663_060 |
0.1022661204 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |