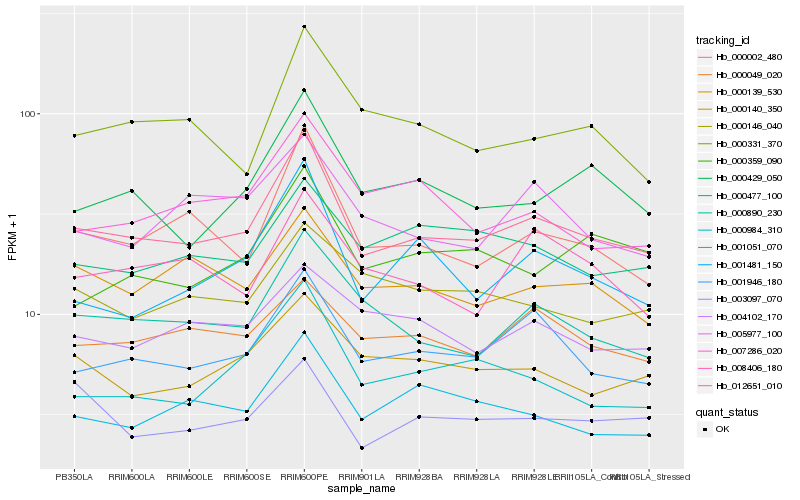
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012651_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 2 |
Hb_001946_180 |
0.0804376654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000002_480 |
0.0851836222 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000890_230 |
0.0893806008 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 5 |
Hb_000429_050 |
0.0959168159 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000140_350 |
0.0982218336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000984_310 |
0.1001794914 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 8 |
Hb_001051_070 |
0.1005033054 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000139_530 |
0.1008791333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000049_020 |
0.1047500149 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 11 |
Hb_001481_150 |
0.1056972047 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 12 |
Hb_000331_370 |
0.1064941791 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 13 |
Hb_005977_100 |
0.1069276947 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 14 |
Hb_007286_020 |
0.1075794212 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 15 |
Hb_000477_100 |
0.1094256213 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 16 |
Hb_008406_180 |
0.1095349764 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_003097_070 |
0.1101612182 |
- |
- |
glucose regulated repressor protein, putative [Ricinus communis] |
| 18 |
Hb_000146_040 |
0.1107281114 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 19 |
Hb_004102_170 |
0.111320418 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 20 |
Hb_000359_090 |
0.111425367 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |