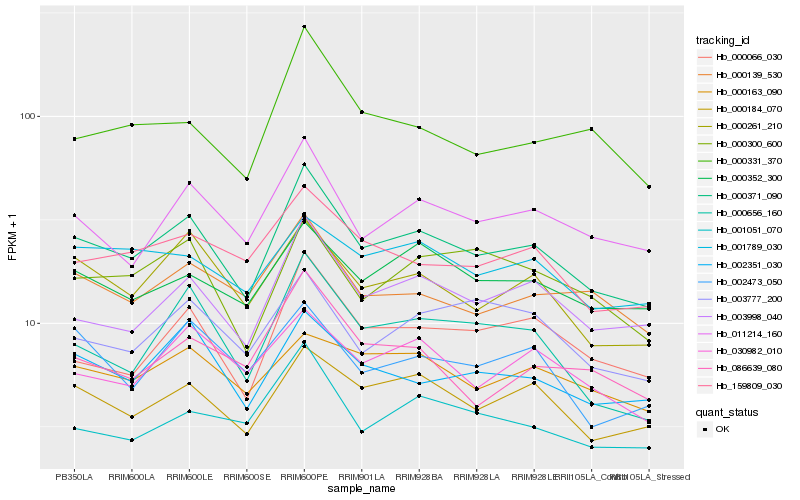
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000371_090 |
0.0 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 2 |
Hb_003998_040 |
0.0731123082 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 3 |
Hb_000184_070 |
0.073671101 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011214_160 |
0.0771254679 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 5 |
Hb_000261_210 |
0.0784089616 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 6 |
Hb_002351_030 |
0.0833362908 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_000066_030 |
0.0835227495 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000656_160 |
0.0859552956 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 9 |
Hb_159809_030 |
0.0882272732 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000139_530 |
0.0895292486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000352_300 |
0.0904779808 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 12 |
Hb_000300_600 |
0.0922412559 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 13 |
Hb_030982_010 |
0.0927621564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001051_070 |
0.0945695893 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003777_200 |
0.0956659475 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 16 |
Hb_002473_050 |
0.0973582063 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_001789_030 |
0.0973822809 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 18 |
Hb_000331_370 |
0.0980391799 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 19 |
Hb_000163_090 |
0.098739119 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 20 |
Hb_086639_080 |
0.099194258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |