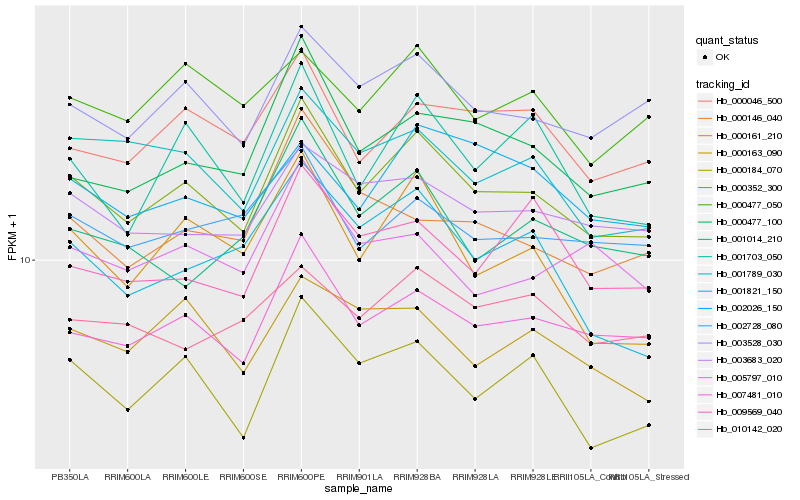
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_300 |
0.0 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 2 |
Hb_007481_010 |
0.0514942593 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 3 |
Hb_003528_030 |
0.0580126803 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 4 |
Hb_000184_070 |
0.0648448624 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000477_100 |
0.0653141868 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 6 |
Hb_000046_500 |
0.0687478836 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 7 |
Hb_003683_020 |
0.0690274462 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_001789_030 |
0.0695786502 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 9 |
Hb_001703_050 |
0.0704692083 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 10 |
Hb_002728_080 |
0.0730736583 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_000163_090 |
0.0757921001 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 12 |
Hb_000161_210 |
0.0770926489 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 13 |
Hb_000477_050 |
0.0771254117 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 14 |
Hb_002026_150 |
0.0790478023 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair helicase XPB1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005797_010 |
0.0818365013 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_000146_040 |
0.081839217 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 17 |
Hb_010142_020 |
0.0824967525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001014_210 |
0.0825425402 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 19 |
Hb_001821_150 |
0.0828767262 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 20 |
Hb_009569_040 |
0.0830559864 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |