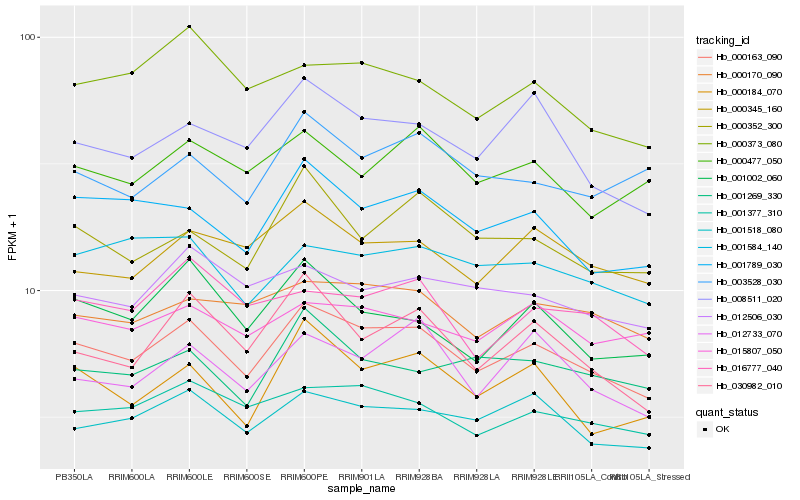
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000163_090 |
0.0 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 2 |
Hb_001518_080 |
0.0653135498 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 3 |
Hb_001789_030 |
0.0656751738 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 4 |
Hb_001377_310 |
0.0680043524 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 5 |
Hb_008511_020 |
0.0713805702 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 6 |
Hb_000184_070 |
0.0728992866 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 7 |
Hb_030982_010 |
0.0732744518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000170_090 |
0.0735585181 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 9 |
Hb_000345_160 |
0.0735789199 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 10 |
Hb_000352_300 |
0.0757921001 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 11 |
Hb_001002_060 |
0.0767560867 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 12 |
Hb_012733_070 |
0.0780684535 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 13 |
Hb_012506_030 |
0.0793586729 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 14 |
Hb_000373_080 |
0.0794587573 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 15 |
Hb_001584_140 |
0.079843593 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 16 |
Hb_015807_050 |
0.0802183904 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 17 |
Hb_001269_330 |
0.0810032486 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 18 |
Hb_016777_040 |
0.0811084893 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 19 |
Hb_003528_030 |
0.0811819007 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 20 |
Hb_000477_050 |
0.0812973829 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |