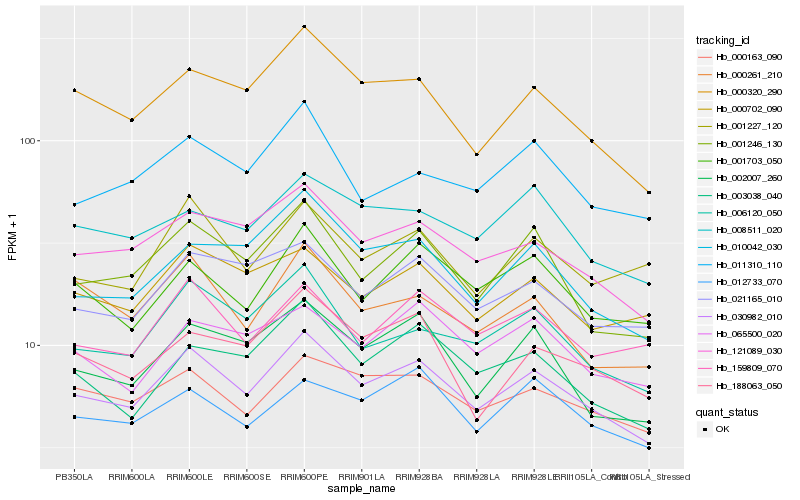
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030982_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_121089_030 |
0.0616871474 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 3 |
Hb_001246_130 |
0.0693233166 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000320_290 |
0.0701090084 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 5 |
Hb_006120_050 |
0.0713388088 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 6 |
Hb_000163_090 |
0.0732744518 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 7 |
Hb_021165_010 |
0.0743988348 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002007_260 |
0.075221479 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 9 |
Hb_001703_050 |
0.0768313286 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 10 |
Hb_000702_090 |
0.0768552747 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 11 |
Hb_003038_040 |
0.0783202668 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 12 |
Hb_159809_070 |
0.0794158296 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 13 |
Hb_008511_020 |
0.0803421577 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 14 |
Hb_010042_030 |
0.0806235366 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 15 |
Hb_065500_020 |
0.0816293748 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 16 |
Hb_012733_070 |
0.0830984409 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001227_120 |
0.0833828542 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 18 |
Hb_011310_110 |
0.0838585367 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000261_210 |
0.0842077304 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 20 |
Hb_188063_050 |
0.084249535 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |