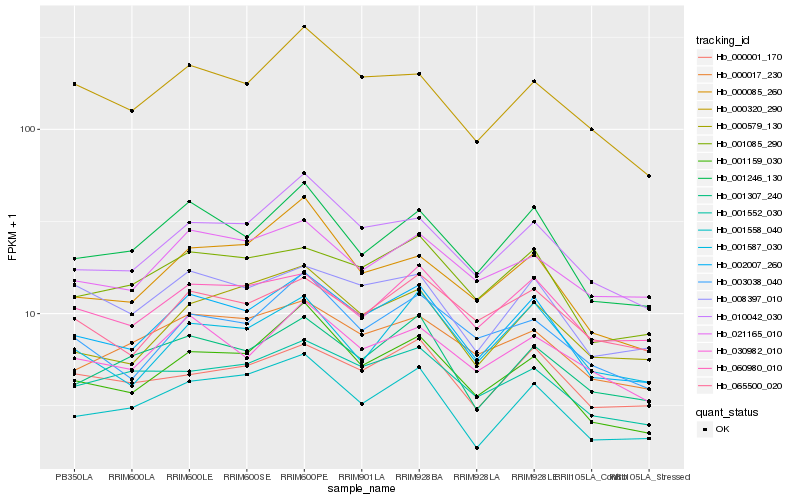
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002007_260 |
0.0 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 2 |
Hb_001246_130 |
0.0562129409 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 3 |
Hb_010042_030 |
0.065609136 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 4 |
Hb_001085_290 |
0.0656987374 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 5 |
Hb_001159_030 |
0.0658225757 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 6 |
Hb_001558_040 |
0.0688906236 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 7 |
Hb_030982_010 |
0.075221479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008397_010 |
0.075747827 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003038_040 |
0.0772658186 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 10 |
Hb_001307_240 |
0.0776637014 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_060980_010 |
0.0780082554 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 12 |
Hb_000579_130 |
0.0797594962 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000320_290 |
0.0801865422 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 14 |
Hb_000017_230 |
0.0803106204 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 15 |
Hb_021165_010 |
0.081883685 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_065500_020 |
0.0830642601 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 17 |
Hb_001552_030 |
0.0839803441 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001587_030 |
0.0846062855 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 19 |
Hb_000085_260 |
0.0846565917 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 20 |
Hb_000001_170 |
0.0848125982 |
- |
- |
amino acid transporter, putative [Ricinus communis] |