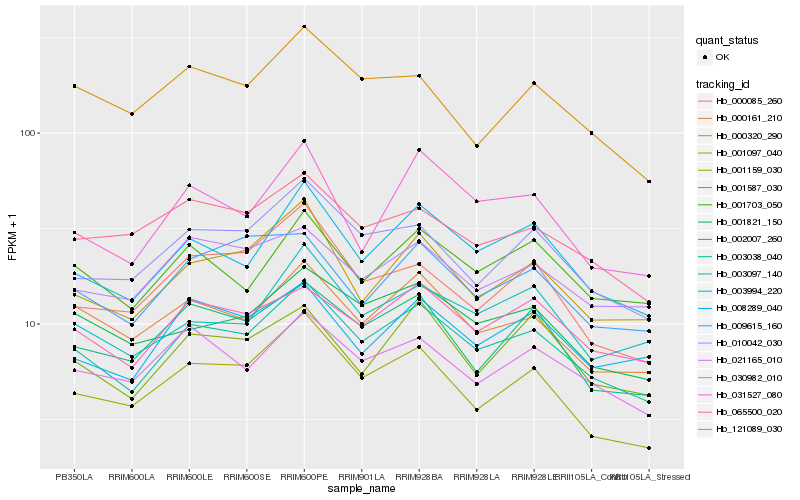
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_040 |
0.0 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 2 |
Hb_003994_220 |
0.0573457455 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 3 |
Hb_000161_210 |
0.0680001328 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 4 |
Hb_001097_040 |
0.0707941911 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 5 |
Hb_065500_020 |
0.0712793484 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 6 |
Hb_001159_030 |
0.0733575886 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 7 |
Hb_010042_030 |
0.0737536137 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 8 |
Hb_001821_150 |
0.0747809281 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 9 |
Hb_001703_050 |
0.0766473967 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 10 |
Hb_002007_260 |
0.0772658186 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 11 |
Hb_030982_010 |
0.0783202668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003097_140 |
0.0829495723 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 13 |
Hb_031527_080 |
0.0833356945 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 14 |
Hb_008289_040 |
0.0835796742 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 15 |
Hb_021165_010 |
0.0845309996 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001587_030 |
0.0867514252 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 17 |
Hb_121089_030 |
0.0871740985 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 18 |
Hb_000085_260 |
0.0900786872 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 19 |
Hb_009615_160 |
0.0903414167 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 20 |
Hb_000320_290 |
0.0923307109 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |