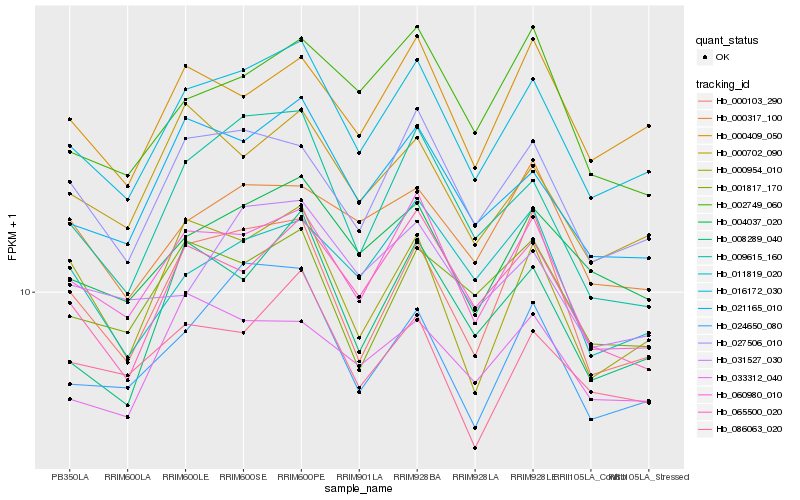
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016172_030 |
0.0 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 2 |
Hb_060980_010 |
0.0433949501 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_021165_010 |
0.0496392605 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004037_020 |
0.0497815828 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 5 |
Hb_027506_010 |
0.0512219878 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 6 |
Hb_065500_020 |
0.0574570188 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 7 |
Hb_024650_080 |
0.0587747648 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 8 |
Hb_000409_050 |
0.0599066516 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 9 |
Hb_000103_290 |
0.0601216449 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 10 |
Hb_008289_040 |
0.0601858779 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 11 |
Hb_000702_090 |
0.0616635273 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 12 |
Hb_009615_160 |
0.0628535657 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 13 |
Hb_086063_020 |
0.0649938356 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 14 |
Hb_033312_040 |
0.0670548295 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_031527_030 |
0.0673356344 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 16 |
Hb_000317_100 |
0.0682227877 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 17 |
Hb_011819_020 |
0.068498295 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000954_010 |
0.0685718193 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 19 |
Hb_001817_170 |
0.0693858465 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 20 |
Hb_002749_060 |
0.0701225123 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |