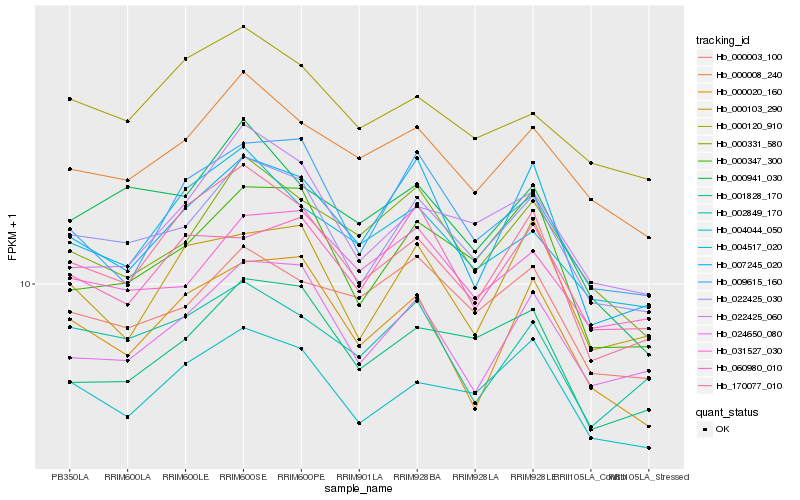
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022425_030 |
0.0 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 2 |
Hb_004044_050 |
0.0574869358 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 3 |
Hb_000347_300 |
0.0610127148 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 4 |
Hb_000008_240 |
0.0630006337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_024650_080 |
0.0638010212 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 6 |
Hb_031527_030 |
0.0695937372 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 7 |
Hb_001828_170 |
0.0696818541 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 8 |
Hb_000331_580 |
0.0701937126 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 9 |
Hb_022425_060 |
0.0713017243 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 10 |
Hb_170077_010 |
0.0719655664 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000003_100 |
0.0744358251 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 12 |
Hb_007245_020 |
0.0774766575 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 13 |
Hb_009615_160 |
0.0781217887 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 14 |
Hb_002849_170 |
0.0795310179 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000120_910 |
0.0796777425 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 16 |
Hb_004517_020 |
0.080366987 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 17 |
Hb_000103_290 |
0.0804746717 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 18 |
Hb_000941_030 |
0.0812047521 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 19 |
Hb_000020_160 |
0.0814884232 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 20 |
Hb_060980_010 |
0.0836900219 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |