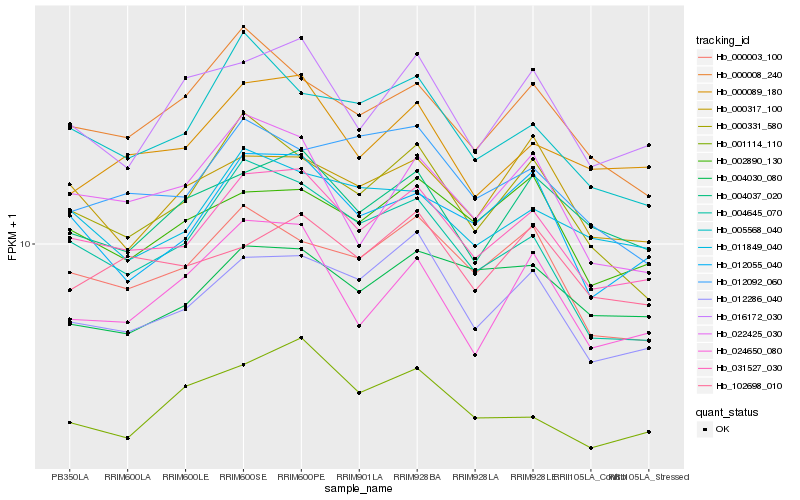
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031527_030 |
0.0 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 2 |
Hb_012286_040 |
0.0534003067 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 3 |
Hb_012055_040 |
0.0615932931 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_000003_100 |
0.0617177746 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 5 |
Hb_004037_020 |
0.0644299909 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000089_180 |
0.0645228118 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 7 |
Hb_011849_040 |
0.0649070878 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_002890_130 |
0.066038725 |
- |
- |
tip120, putative [Ricinus communis] |
| 9 |
Hb_016172_030 |
0.0673356344 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 10 |
Hb_102698_010 |
0.0673788483 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000331_580 |
0.0678068268 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 12 |
Hb_000008_240 |
0.067906565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004645_070 |
0.0689515623 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 14 |
Hb_005568_040 |
0.0692192652 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 15 |
Hb_022425_030 |
0.0695937372 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 16 |
Hb_001114_110 |
0.07187997 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 17 |
Hb_012092_060 |
0.0731143212 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 18 |
Hb_024650_080 |
0.0732886591 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 19 |
Hb_000317_100 |
0.0734546774 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 20 |
Hb_004030_080 |
0.0745528722 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |