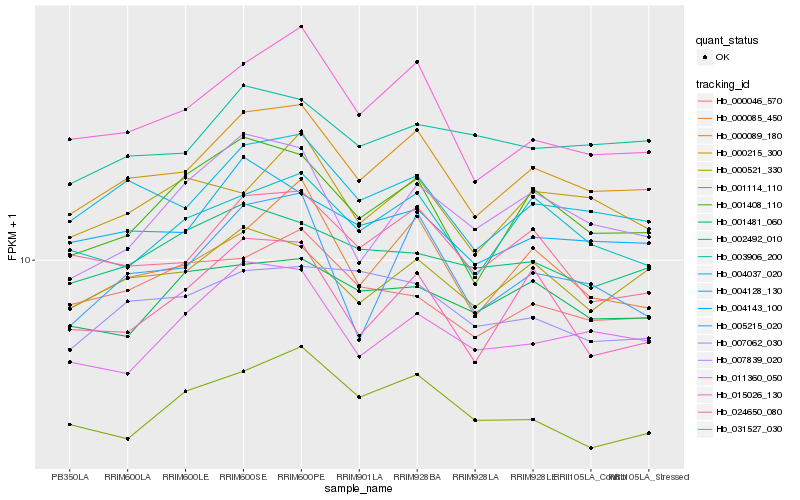
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000089_180 |
0.0 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 2 |
Hb_004128_130 |
0.0434889389 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 3 |
Hb_015026_130 |
0.0577157069 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 4 |
Hb_000085_450 |
0.05874205 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 5 |
Hb_004143_100 |
0.0605972096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001408_110 |
0.0609543131 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 7 |
Hb_004037_020 |
0.0638202085 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 8 |
Hb_031527_030 |
0.0645228118 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 9 |
Hb_000521_330 |
0.0647451422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011360_050 |
0.0709639576 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003906_200 |
0.0729797259 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 12 |
Hb_000215_300 |
0.0748575356 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 13 |
Hb_024650_080 |
0.0754317065 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 14 |
Hb_005215_020 |
0.0762244332 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 15 |
Hb_007839_020 |
0.077878019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002492_010 |
0.0780503297 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 17 |
Hb_001114_110 |
0.0785623922 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 18 |
Hb_000046_570 |
0.0788216295 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 19 |
Hb_007062_030 |
0.0792803261 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 20 |
Hb_001481_060 |
0.0798988493 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |