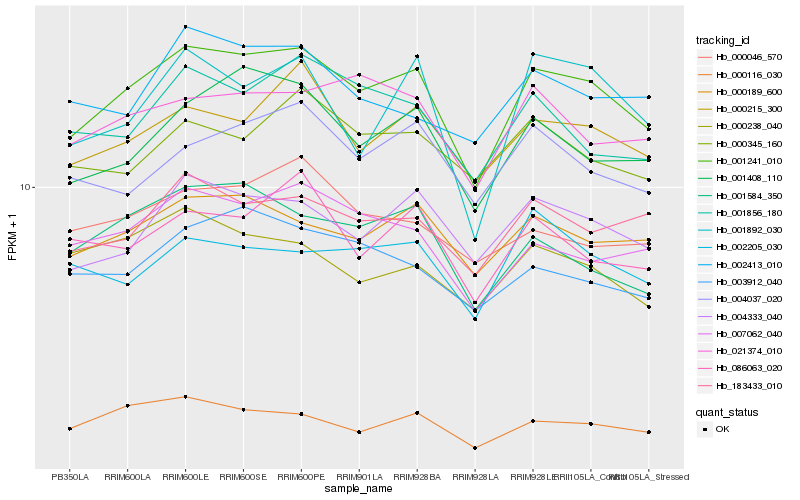
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001241_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_004333_040 |
0.0680267024 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 3 |
Hb_000189_600 |
0.070376912 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_001408_110 |
0.072091316 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 5 |
Hb_000215_300 |
0.0729755748 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 6 |
Hb_183433_010 |
0.0746143282 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 7 |
Hb_000116_030 |
0.0761829872 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 8 |
Hb_007062_040 |
0.0767425383 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 9 |
Hb_021374_010 |
0.081107521 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 10 |
Hb_001856_180 |
0.0816558808 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 11 |
Hb_001584_350 |
0.0819769753 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 12 |
Hb_086063_020 |
0.0827191701 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 13 |
Hb_004037_020 |
0.082807086 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003912_040 |
0.0847337475 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 15 |
Hb_000345_160 |
0.0853935287 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 16 |
Hb_001892_030 |
0.0882904318 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase isoform X1 [Jatropha curcas] |
| 17 |
Hb_000046_570 |
0.0884955294 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 18 |
Hb_000238_040 |
0.0890577116 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 19 |
Hb_002413_010 |
0.0891138189 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_002205_030 |
0.0902877275 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |