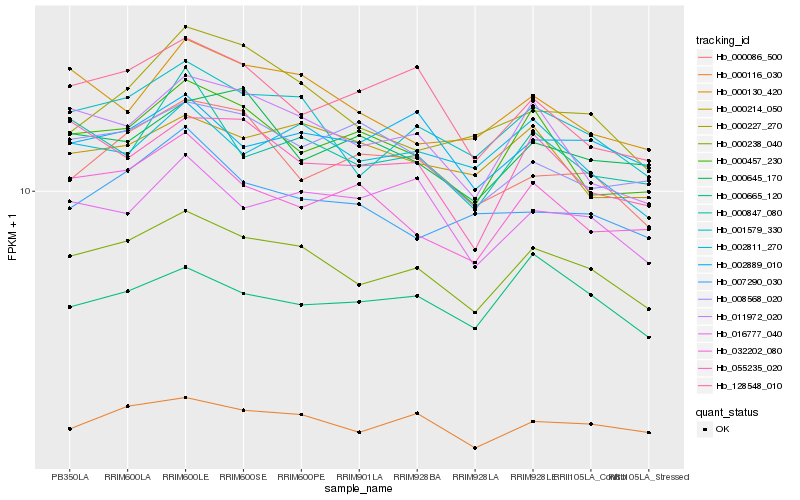
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000238_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 2 |
Hb_001579_330 |
0.0364345777 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_011972_020 |
0.0607293165 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_000116_030 |
0.0642927966 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 5 |
Hb_002811_270 |
0.0721249706 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 6 |
Hb_000130_420 |
0.0742023325 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 7 |
Hb_000457_230 |
0.0744871184 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_055235_020 |
0.0746849488 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002889_010 |
0.0763155819 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 10 |
Hb_000214_050 |
0.0777183417 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000847_080 |
0.0777688929 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 12 |
Hb_016777_040 |
0.0787907393 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 13 |
Hb_032202_080 |
0.0789444783 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000665_120 |
0.0796063116 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 15 |
Hb_000086_500 |
0.0796289391 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_007290_030 |
0.0805752495 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 17 |
Hb_128548_010 |
0.081308787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008568_020 |
0.0820709308 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 19 |
Hb_000645_170 |
0.0827431831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000227_270 |
0.0827932682 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |