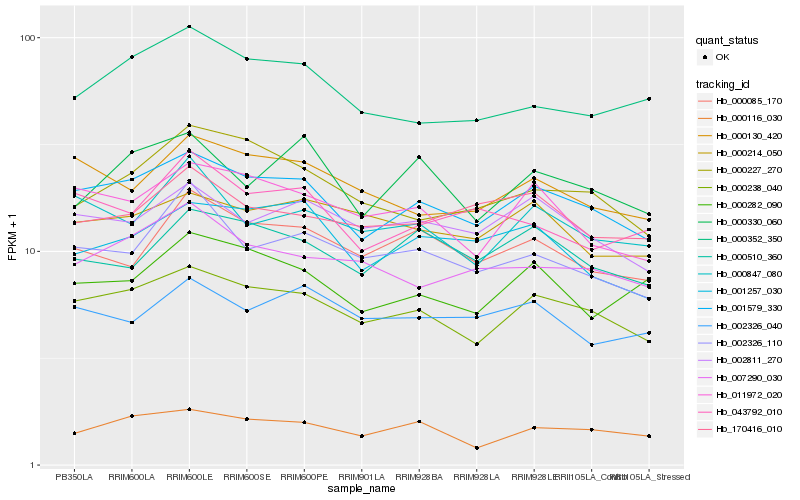
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001579_330 |
0.0 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000238_040 |
0.0364345777 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 3 |
Hb_002811_270 |
0.0723053723 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 4 |
Hb_011972_020 |
0.0732776015 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000130_420 |
0.0782233913 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 6 |
Hb_001257_030 |
0.0798661005 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 7 |
Hb_000116_030 |
0.0803167484 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 8 |
Hb_000510_360 |
0.0813848895 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 9 |
Hb_000085_170 |
0.0839202216 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007290_030 |
0.0849164131 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 11 |
Hb_000227_270 |
0.085002511 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000847_080 |
0.0850103299 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 13 |
Hb_000282_090 |
0.0854934775 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 14 |
Hb_002326_110 |
0.086148484 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 15 |
Hb_000214_050 |
0.0863328083 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_043792_010 |
0.0867295051 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000352_350 |
0.0867316702 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 18 |
Hb_170416_010 |
0.0878939837 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 19 |
Hb_000330_060 |
0.0879350158 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 20 |
Hb_002326_040 |
0.0890436214 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |