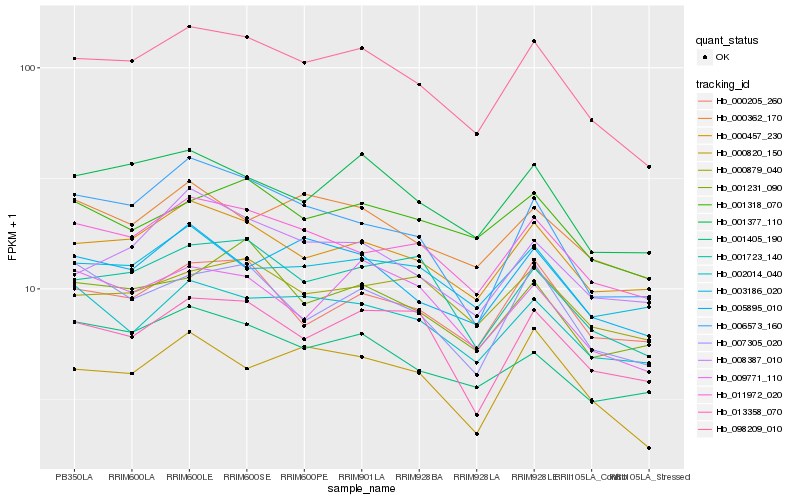
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098209_010 |
0.0 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 2 |
Hb_000457_230 |
0.0613962935 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_011972_020 |
0.0614072484 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_001377_110 |
0.0629997191 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 5 |
Hb_001723_140 |
0.0691000121 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_005895_010 |
0.0703531727 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002014_040 |
0.0732533898 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 8 |
Hb_009771_110 |
0.0752046891 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 9 |
Hb_000205_260 |
0.0757689547 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 10 |
Hb_007305_020 |
0.0761282687 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 11 |
Hb_000362_170 |
0.076364315 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 12 |
Hb_001405_190 |
0.0764648839 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_013358_070 |
0.0783380775 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_001318_070 |
0.0786212886 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003186_020 |
0.0794494331 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006573_160 |
0.080260774 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 17 |
Hb_000820_150 |
0.080334757 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 18 |
Hb_008387_010 |
0.0805257173 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 19 |
Hb_000879_040 |
0.080623641 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 20 |
Hb_001231_090 |
0.0812537556 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |