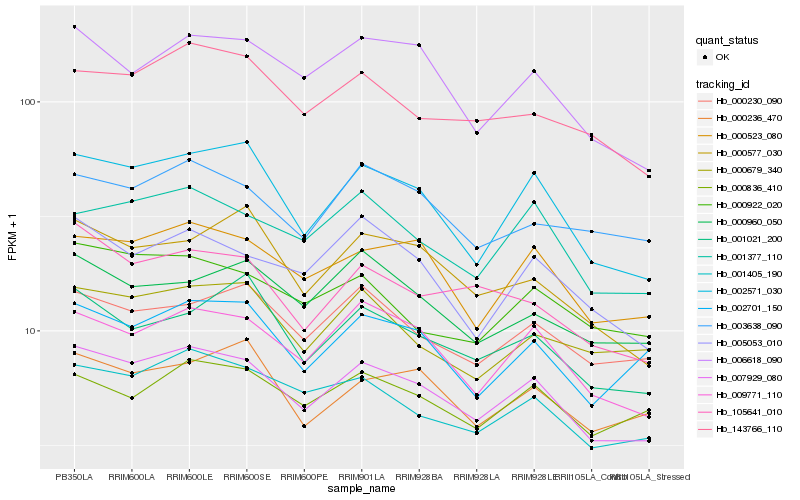
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007929_080 |
0.0 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 2 |
Hb_002571_030 |
0.0523524771 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 3 |
Hb_009771_110 |
0.0544382314 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 4 |
Hb_000523_080 |
0.0578480331 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 5 |
Hb_001405_190 |
0.06026951 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001377_110 |
0.0649091769 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 7 |
Hb_006618_090 |
0.0676909571 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 8 |
Hb_143766_110 |
0.0704396119 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 9 |
Hb_000230_090 |
0.070509246 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005053_010 |
0.0727713736 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 11 |
Hb_002701_150 |
0.0729081046 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001021_200 |
0.0750702006 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 13 |
Hb_000922_020 |
0.0752623344 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_105641_010 |
0.0757977656 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 15 |
Hb_003638_090 |
0.0768115154 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 16 |
Hb_000236_470 |
0.0777497546 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 17 |
Hb_000679_340 |
0.0777834471 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000960_050 |
0.078357757 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 19 |
Hb_000836_410 |
0.078398636 |
- |
- |
sec10, putative [Ricinus communis] |
| 20 |
Hb_000577_030 |
0.0784156072 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |