| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
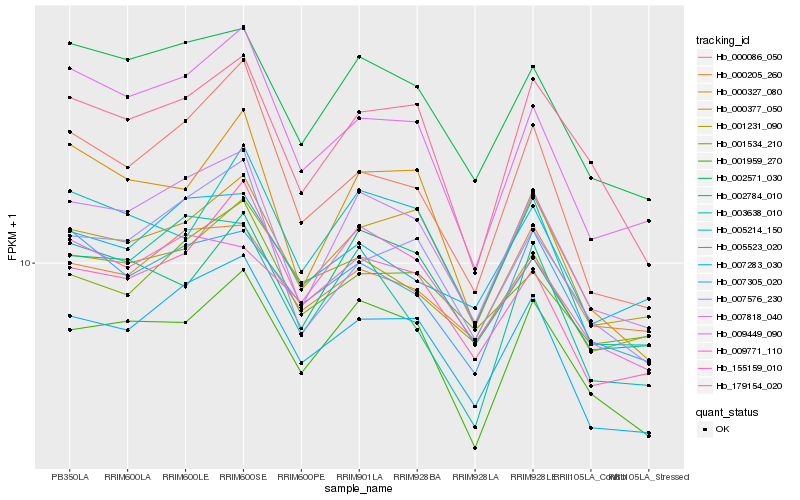
Hb_007818_040 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002571_030 |
0.0503739434 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 3 |
Hb_007283_030 |
0.0610188736 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 4 |
Hb_007576_230 |
0.0632947005 |
- |
- |
hypothetical protein JCGZ_25616 [Jatropha curcas] |
| 5 |
Hb_001959_270 |
0.0686983491 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_179154_020 |
0.0729637524 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_000377_050 |
0.0731216454 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 8 |
Hb_009449_090 |
0.0779526785 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 9 |
Hb_155159_010 |
0.0821873119 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 10 |
Hb_007305_020 |
0.0851438713 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 11 |
Hb_005214_150 |
0.0853107664 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 12 |
Hb_000086_050 |
0.0870163484 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 13 |
Hb_002784_010 |
0.0876680947 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 14 |
Hb_000205_260 |
0.0883850762 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 15 |
Hb_003638_010 |
0.0897811913 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 16 |
Hb_001534_210 |
0.0902180643 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 17 |
Hb_009771_110 |
0.0904194773 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 18 |
Hb_001231_090 |
0.0905914736 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000327_080 |
0.0911110543 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 20 |
Hb_005523_020 |
0.0919864665 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |