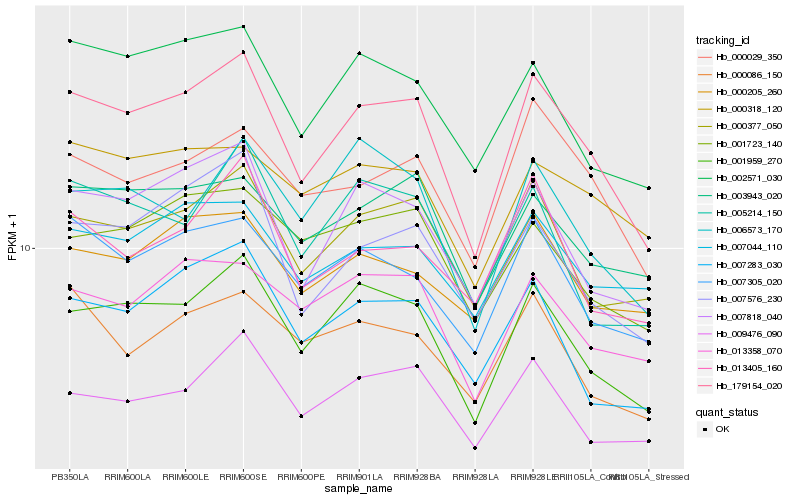
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_179154_020 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_001959_270 |
0.0590709484 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_007818_040 |
0.0729637524 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000377_050 |
0.0774453981 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 5 |
Hb_007305_020 |
0.0840688107 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 6 |
Hb_007576_230 |
0.084762413 |
- |
- |
hypothetical protein JCGZ_25616 [Jatropha curcas] |
| 7 |
Hb_002571_030 |
0.0851917715 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 8 |
Hb_000029_350 |
0.0895961327 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 9 |
Hb_009476_090 |
0.0905088695 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_013358_070 |
0.0907925678 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_007283_030 |
0.0911915705 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 12 |
Hb_003943_020 |
0.0913879001 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007044_110 |
0.0974138778 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_001723_140 |
0.0979108469 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_000318_120 |
0.0986549245 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000086_150 |
0.0987728771 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006573_170 |
0.0993270363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005214_150 |
0.0993355789 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 19 |
Hb_000205_260 |
0.0999383547 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 20 |
Hb_013405_160 |
0.1004063998 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |