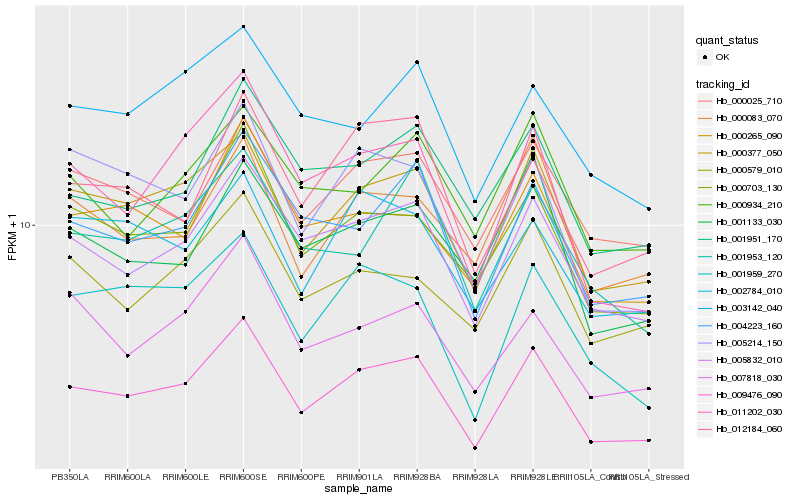
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009476_090 |
0.0 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_000377_050 |
0.0412697996 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 3 |
Hb_005832_010 |
0.0473056435 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000703_130 |
0.0604622579 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 5 |
Hb_004223_160 |
0.0633177236 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000025_710 |
0.0706543742 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 7 |
Hb_001959_270 |
0.0735710115 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_000265_090 |
0.0739363569 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 9 |
Hb_001951_170 |
0.0749362048 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 10 |
Hb_001953_120 |
0.076138265 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011202_030 |
0.0763692082 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 12 |
Hb_000579_010 |
0.0764694342 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 13 |
Hb_007818_030 |
0.0771119239 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 14 |
Hb_000083_070 |
0.0775022662 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_012184_060 |
0.0777397765 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005214_150 |
0.0784296765 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 17 |
Hb_002784_010 |
0.0793998533 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 18 |
Hb_001133_030 |
0.0796467973 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 19 |
Hb_003142_040 |
0.0797199543 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 20 |
Hb_000934_210 |
0.0801027199 |
- |
- |
suppressor of ty, putative [Ricinus communis] |