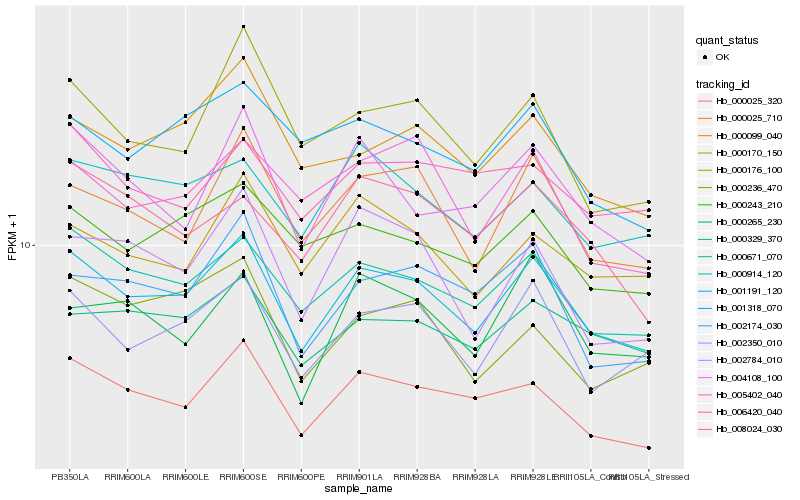
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001191_120 |
0.0 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000671_070 |
0.0447954388 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 3 |
Hb_000025_320 |
0.0483540456 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 4 |
Hb_002350_010 |
0.0527834523 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 5 |
Hb_000025_710 |
0.0616783509 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 6 |
Hb_000243_210 |
0.0640084662 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_000914_120 |
0.0643302849 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000329_370 |
0.0664079901 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 9 |
Hb_000099_040 |
0.0664476028 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_000170_150 |
0.0666987407 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 11 |
Hb_004108_100 |
0.0678131751 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 12 |
Hb_002174_030 |
0.0684418713 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 13 |
Hb_000176_100 |
0.0714801698 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 14 |
Hb_001318_070 |
0.0720022774 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002784_010 |
0.0727242203 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 16 |
Hb_008024_030 |
0.0736523666 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 17 |
Hb_005402_040 |
0.0737527673 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 18 |
Hb_006420_040 |
0.0739897876 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000265_230 |
0.0746162939 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105643603 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000236_470 |
0.0759194887 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |